My source data looks like this:
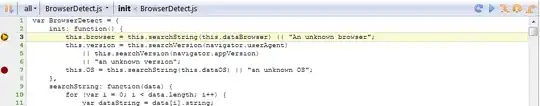
data <- list(row1 = list(D = data.table(stat = "D", est = 0.2), HM = data.table(stat = c("H","M"), est = c(0.4,0.5))),
row2 = list(D = data.table(stat = "D", est = 0.3), HM = data.table(stat = c("H","M"), est = c(0.1,0.6))))
I want it to look like this:
I apply some solutions in this post, for de-nesting lists, for example:
as.data.frame(t(sapply(data, unlist)))
But I get:
Seems like I could do one more step and try to manually clean the latter, but the actual data is more populous and with more variables. I thought I needed to first reconvert doubles (vectors) into singles but after that result is the same. Any ideas?