I'm trying to analyze some noisy time-series data in R. The data are based on the CO2 emission of animals and they show a sort of cyclic periodicity that I'd like to characterize. I'd like to test the hypotheses:
H0: There is no cyclic CO2 emission (i.e. no more than random).
H1: There is a pattern of CO2 emission in cycles or pulses.
So to do this I've imported the data into R, converted it to a time series class, and plotted its periodogram.
t25a <- read.table("data.txt", header=TRUE, sep="\t")
t1 <- ts(t25a$Co2)
plot(t1)
spec.pgram(t1, spans=4, log="no")
Here's what that looks like, with the raw data plotted on top and the periodogram beneath:
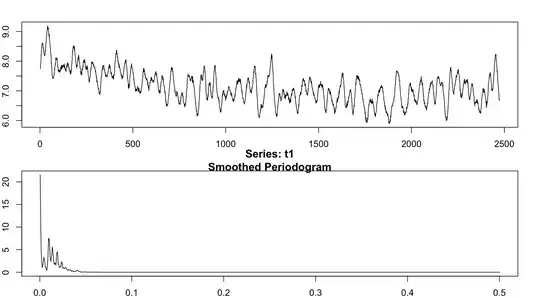
In the bottom figure, I can see four or five somewhat-distinct peaks indicating a frequency component in the data. My question is -- are they all equally "important"? Is there any way to test whether the observed peaks are significantly different from each other or from the predictions of the null hypothesis? All I know how to do is find the frequency associated with those peaks, but I'd like a more objective method for determining how many "significant" peaks there really are in the data.