I developed new algorithm for phylogenetic tree comparison(phylogenetic tree is simply rooted binary tree). As an input we have two trees, we want to calculate their similarity percentage. one example of these type of algorithms is here.
But most of these algorithms(as I know all of them) did not offer a good way to check the accuracy of their algorithms. e.g if you look at the following figure, you can see there is more similarity between T1 and T3, versus T1 and T2.
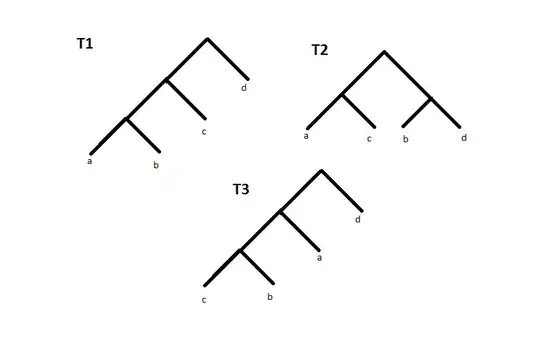
I need a method for checking its accuracy of similarity measure, To be sure that my algorithm is better than previous algorithms !!! (it is not difficult in most of the case by human eyes but I don't know how to extend it to my application)
your validity measure should be independent from algorithm.