How can I create a cluster plot in R without using clustplot?
I am trying to get to grips with some clustering (using R) and visualisation (using HTML5 Canvas).
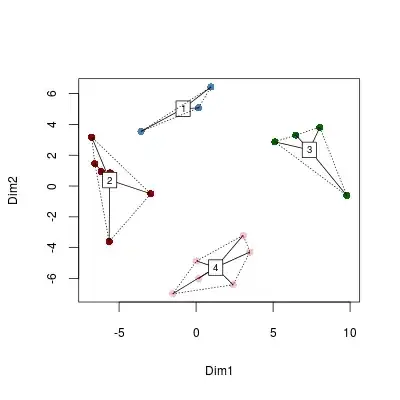
Basically, I want to create a cluster plot but instead of plotting the data, I want to get a set of 2D points or coordinates that I can pull into canvas and do something might pretty with (but I am unsure of how to do this). I would imagine that I:
- Create a similarity matrix for the entire dataset (using dist)
- Cluster the similarity matrix using kmeans or something similar (using kmeans)
- Plot the result using MDS or PCA - but I am unsure of how steps 2 and 3 relate (cmdscale).
I've checked out questions here, here and here (with the last one being of most use).