A fast numpy only method is to pad the array (if you want to consider edges), then compare shifted slices. See localMax function below.
import numpy as np
import matplotlib.pyplot as plt
# Generate cloudy noise
Nx,Ny = 100,200
grid = np.mgrid[-1:1:1j*Nx,-1:1:1j*Ny]
filt = 10**(-10*(1-grid**2).mean(axis=0)) # Low pass filter
cloudy = np.abs(np.fft.ifft2((np.random.rand(Nx,Ny)-.5)*filt))/filt.mean()
# Generate checkerboard on a large peak
Nx,Ny = 10,20
checkerboard = 1.0*np.mgrid[0:Nx,0:Ny].sum(axis=0)%2
checkerboard *= 2-(np.mgrid[-1:1:1j*Nx,-1:1:1j*Ny]**2).sum(axis=0)
def localMax(a, include_diagonal=True, threshold=-np.inf) :
# Pad array so we can handle edges
ap = np.pad(a, ((1,1),(1,1)), constant_values=-np.inf )
# Determines if each location is bigger than adjacent neighbors
adjacentmax =(
(ap[1:-1,1:-1] > threshold) &
(ap[0:-2,1:-1] <= ap[1:-1,1:-1]) &
(ap[2:, 1:-1] <= ap[1:-1,1:-1]) &
(ap[1:-1,0:-2] <= ap[1:-1,1:-1]) &
(ap[1:-1,2: ] <= ap[1:-1,1:-1])
)
if not include_diagonal :
return np.argwhere(adjacentmax)
# Determines if each location is bigger than diagonal neighbors
diagonalmax =(
(ap[0:-2,0:-2] <= ap[1:-1,1:-1]) &
(ap[2: ,2: ] <= ap[1:-1,1:-1]) &
(ap[0:-2,2: ] <= ap[1:-1,1:-1]) &
(ap[2: ,0:-2] <= ap[1:-1,1:-1])
)
return np.argwhere(adjacentmax & diagonalmax)
plt.figure(1); plt.clf()
plt.imshow(cloudy, cmap='bone')
mx1 = localMax(cloudy)
#mx1 = np.argwhere(maximum_filter(cloudy, size=3)==cloudy) # Compare scipy filter
mx2 = localMax(cloudy, threshold=cloudy.mean()*.8)
plt.scatter(mx1[:,1],mx1[:,0], color='yellow', s=20)
plt.scatter(mx2[:,1],mx2[:,0], color='red', s=5)
plt.savefig('localMax1.png')
plt.figure(2); plt.clf()
plt.imshow(checkerboard, cmap='bone')
mx1 = localMax(checkerboard,False)
mx2 = localMax(checkerboard)
plt.scatter(mx1[:,1],mx1[:,0], color='yellow', s=20)
plt.scatter(mx2[:,1],mx2[:,0], color='red', s=10)
plt.savefig('localMax2.png')
plt.show()
Time is about the same with the scipy filter:
In [169]: %timeit mx2 = np.argwhere((maximum_filter(cloudy, size=3)==cloudy) & (cloudy>.5))
244 µs ± 1.1 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
In [172]: %timeit mx1 = localMax(cloudy, True, .5)
262 µs ± 1.44 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
Here's the 'cloudy' example with all peaks (yellow) and above threshold peaks (red):
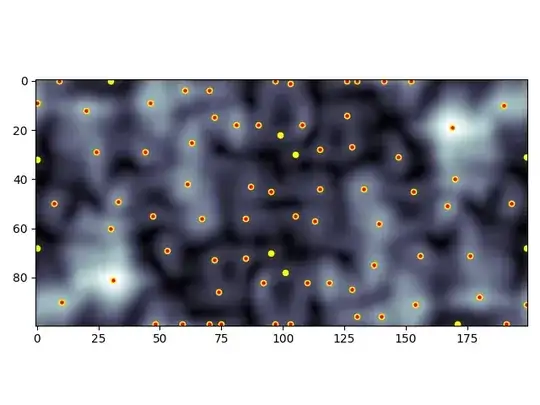
Whether or not you include diagonals could be important depending on your use case. The checkerboard demonstrates that, with peaks not considering diagonals (yellow) and peaks that do consider diagonals (red):