I use the R package GBM as probably my first choice for predictive modeling. There are so many great things about this algorithm but the one "bad" is that I cant easily use model code to score new data outside of R. I want to write code that can be used in SAS or other system (I will start with SAS (no access to IML)).
Lets say I have the following data set (from GBM manual) and model code:
library(gbm)
set.seed(1234)
N <- 1000
X1 <- runif(N)
X2 <- 2*runif(N)
X3 <- ordered(sample(letters[1:4],N,replace=TRUE),levels=letters[4:1])
X4 <- factor(sample(letters[1:6],N,replace=TRUE))
X5 <- factor(sample(letters[1:3],N,replace=TRUE))
X6 <- 3*runif(N)
mu <- c(-1,0,1,2)[as.numeric(X3)]
SNR <- 10 # signal-to-noise ratio
Y <- X1**1.5 + 2 * (X2**.5) + mu
sigma <- sqrt(var(Y)/SNR)
Y <- Y + rnorm(N,0,sigma)
# introduce some missing values
#X1[sample(1:N,size=500)] <- NA
X4[sample(1:N,size=300)] <- NA
X3[sample(1:N,size=30)] <- NA
data <- data.frame(Y=Y,X1=X1,X2=X2,X3=X3,X4=X4,X5=X5,X6=X6)
# fit initial model
gbm1 <- gbm(Y~X1+X2+X3+X4+X5+X6, # formula
data=data, # dataset
var.monotone=c(0,0,0,0,0,0), # -1: monotone decrease,
distribution="gaussian",
n.trees=2, # number of trees
shrinkage=0.005, # shrinkage or learning rate,
# 0.001 to 0.1 usually work
interaction.depth=5, # 1: additive model, 2: two-way interactions, etc.
bag.fraction = 1, # subsampling fraction, 0.5 is probably best
train.fraction = 1, # fraction of data for training,
# first train.fraction*N used for training
n.minobsinnode = 10, # minimum total weight needed in each node
cv.folds = 5, # do 5-fold cross-validation
keep.data=TRUE, # keep a copy of the dataset with the object
verbose=TRUE) # print out progress
Now I can see the individual trees using pretty.gbm.tree as in
pretty.gbm.tree(gbm1,i.tree = 1)[1:7]
which yields
SplitVar SplitCodePred LeftNode RightNode MissingNode ErrorReduction Weight
0 2 1.5000000000 1 8 15 983.34315 1000
1 1 1.0309565491 2 6 7 190.62220 501
2 2 0.5000000000 3 4 5 75.85130 277
3 -1 -0.0102671518 -1 -1 -1 0.00000 139
4 -1 -0.0050342273 -1 -1 -1 0.00000 138
5 -1 -0.0076601353 -1 -1 -1 0.00000 277
6 -1 -0.0014569934 -1 -1 -1 0.00000 224
7 -1 -0.0048866747 -1 -1 -1 0.00000 501
8 1 0.6015416372 9 10 14 160.97007 469
9 -1 0.0007403551 -1 -1 -1 0.00000 142
10 2 2.5000000000 11 12 13 85.54573 327
11 -1 0.0046278704 -1 -1 -1 0.00000 168
12 -1 0.0097445692 -1 -1 -1 0.00000 159
13 -1 0.0071158065 -1 -1 -1 0.00000 327
14 -1 0.0051854993 -1 -1 -1 0.00000 469
15 -1 0.0005408284 -1 -1 -1 0.00000 30
The manual page 18 shows the following:
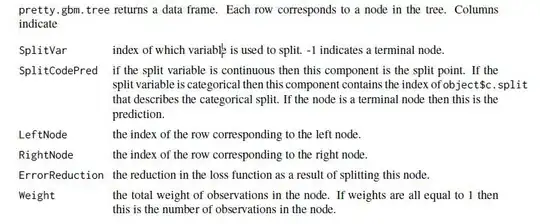
Based on the manual, the first split occurs on the 3rd variable (zero based in this output) which is gbm1$var.names[3] "X3". The variable is ordered factor.
types<-lapply (lapply(data[,gbm1$var.names],class), function(i) ifelse (strsplit(i[1]," ")[1]=="ordered","ordered",i))
types[3]
So, the split is at 1.5 meaning the value 'd and c' levels[[3]][1:2.5] (also zero based) splits to left node and the others levels[[3]][3:4] go to the right.
Next, the rule continues with a split at gbm1$var.names[2] as denoted by SplitVar=1 in the row indexed 1.
Has anyone written anything to move through this data structure (for each tree), constructing rules such as:
"If X3 in ('d','c') and X2<1.0309565491 and X3 in ('d') then scoreTreeOne= -0.0102671518"
which is how I think the first rule from this tree reads.
Or have any advice how to best do this?