I use lme4 in R to fit the mixed model
lmer(value~status+(1|experiment)))
where value is continuous, status(N/D/R) and experiment are factors, and I get
Linear mixed model fit by REML
Formula: value ~ status + (1 | experiment)
AIC BIC logLik deviance REMLdev
29.1 46.98 -9.548 5.911 19.1
Random effects:
Groups Name Variance Std.Dev.
experiment (Intercept) 0.065526 0.25598
Residual 0.053029 0.23028
Number of obs: 264, groups: experiment, 10
Fixed effects:
Estimate Std. Error t value
(Intercept) 2.78004 0.08448 32.91
statusD 0.20493 0.03389 6.05
statusR 0.88690 0.03583 24.76
Correlation of Fixed Effects:
(Intr) statsD
statusD -0.204
statusR -0.193 0.476
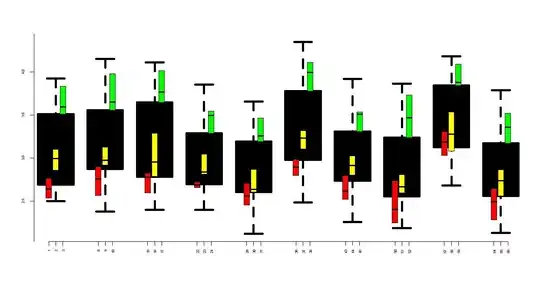
I would like to graphically represent the fixed effects evaluation. However the seems to be no plot function for these objects. Is there any way I can graphically depict the fixed effects?