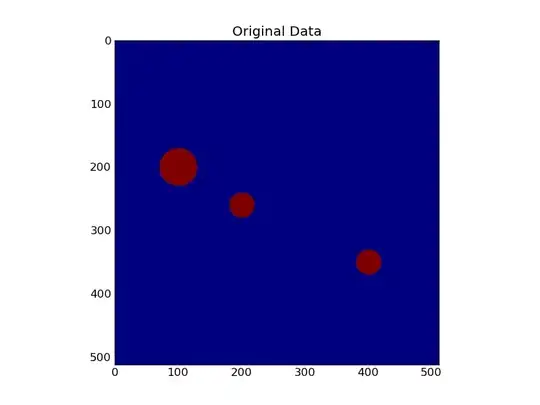
My goal is to trace drawings that have a lot of separate shapes in them and to split these shapes into individual images. It is black on white. I'm quite new to numpy,opencv&co - but here is my current thought:
- scan for black pixels
- black pixel found -> watershed
- find watershed boundary (as polygon path)
- continue searching, but ignore points within the already found boundaries
I'm not very good at these kind of things, is there a better way?
First I tried to find the rectangular bounding box of the watershed results (this is more or less a collage of examples):
from numpy import *
import numpy as np
from scipy import ndimage
np.set_printoptions(threshold=np.nan)
a = np.zeros((512, 512)).astype(np.uint8) #unsigned integer type needed by watershed
y, x = np.ogrid[0:512, 0:512]
m1 = ((y-200)**2 + (x-100)**2 < 30**2)
m2 = ((y-350)**2 + (x-400)**2 < 20**2)
m3 = ((y-260)**2 + (x-200)**2 < 20**2)
a[m1+m2+m3]=1
markers = np.zeros_like(a).astype(int16)
markers[0, 0] = 1
markers[200, 100] = 2
markers[350, 400] = 3
markers[260, 200] = 4
res = ndimage.watershed_ift(a.astype(uint8), markers)
unique(res)
B = argwhere(res.astype(uint8))
(ystart, xstart), (ystop, xstop) = B.min(0), B.max(0) + 1
tr = a[ystart:ystop, xstart:xstop]
print tr
Somehow, when I use the original array (a) then argwhere seems to work, but after the watershed (res) it just outputs the complete array again.
The next step could be to find the polygon path around the shape, but the bounding box would be great for now!
Please help!