DNA adenine methyltransferase identification, often abbreviated DamID,[1] is a molecular biology protocol used to map the binding sites of DNA- and chromatin-binding proteins in eukaryotes. DamID identifies binding sites by expressing the proposed DNA-binding protein as a fusion protein with DNA methyltransferase. Binding of the protein of interest to DNA localizes the methyltransferase in the region of the binding site. Adenine methylation does not occur naturally in eukaryotes and therefore adenine methylation in any region can be concluded to have been caused by the fusion protein, implying the region is located near a binding site. DamID is an alternate method to ChIP-on-chip or ChIP-seq.[2]
Description
Principle
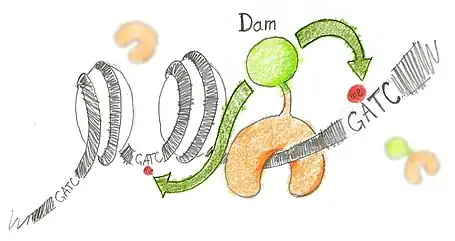
N6-methyladenine (m6A) is the product of the addition of a methyl group (CH3) at position 6 of the adenine. This modified nucleotide is absent from the vast majority of eukaryotes, with the exception of C. elegans,[3] but is widespread in bacterial genomes,[4] as part of the restriction modification or DNA repair systems. In Escherichia coli, adenine methylation is catalyzed by the adenine methyltransferase Dam (DNA adenine methyltransferase), which catalyses adenine methylation exclusively in the palindromic sequence GATC. Ectopic expression of Dam in eukaryotic cells leads to methylation of adenine in GATC sequences without any other noticeable side effect.
Based on this, DamID consists in fusing Dam to a protein of interest (usually a protein that interacts with DNA such as transcription factors) or a chromatin component. The protein of interest thus targets Dam to its cognate in vivo binding site, resulting in the methylation of neighboring GATCs. The presence of m6A, coinciding with the binding sites of the proteins of interest, is revealed by methyl PCR.
Methyl PCR
In this assay the genome is digested by DpnI, which cuts only methylated GATCs. Double-stranded adapters with a known sequence are then ligated to the ends generated by DpnI. Ligation products are then digested by DpnII. This enzyme cuts non-methylated GATCs, ensuring that only fragments flanked by consecutive methylated GATCs are amplified in the subsequent PCR. A PCR with primers matching the adaptors is then carried out, leading to the specific amplification of genomic fragments flanked by methylated GATCs.
Specificities of DamID versus Chromatin Immuno-Precipitation
Chromatin Immuno-Precipitation, or (ChIP), is an alternative method to assay protein binding at specific loci of the genome. Unlike ChIP, DamID does not require a specific antibody against the protein of interest. On the one hand, this allows to map proteins for which no such antibody is available. On the other hand, this makes it impossible to specifically map posttranslationally modified proteins.
Another fundamental difference is that ChIP assays where the protein of interests is at a given time, whereas DamID assays where it has been. The reason is that m6A stays in the DNA after the Dam fusion protein goes away. For proteins that are either bound or unbound on their target sites this does not change the big picture. However, this can lead to strong differences in the case of proteins that slide along the DNA (e.g. RNA polymerase).
Known biases and technical issues
Plasmid methylation bias
Depending on how the experiment is carried out, DamID can be subject to plasmid methylation biases. Because plasmids are usually amplified in E. coli where Dam is naturally expressed, they are methylated on every GATC. In transient transfection experiments, the DNA of those plasmids is recovered along with the DNA of the transfected cells, meaning that fragments of the plasmid are amplified in the methyl PCR. Every sequence of the genome that shares homology or identity with the plasmid may thus appear to be bound by the protein of interest. In particular, this is true of the open reading frame of the protein of interest, which is present in both the plasmid and the genome. In microarray experiments, this bias can be used to ensure that the proper material was hybridized. In stable cell lines or fully transgenic animals, this bias is not observed as no plasmid DNA is recovered.
Apoptosis
Apoptotic cells degrade their DNA in a characteristic nucleosome ladder pattern. This generates DNA fragments that can be ligated and amplified during the DamID procedure (van Steensel laboratory, unpublished observations). The influence of these nucleosomal fragments on the binding profile of a protein is not known.
Resolution
The resolution of DamID is a function of the availability of GATC sequences in the genome. A protein can only be mapped within two consecutive GATC sites. The median spacing between GATC fragments is 205 bp in Drosophila (FlyBase release 5), 260 in mouse (Mm9), and 460 in human (HG19). A modified protocol (DamIP), which combines immunoprecipitation of m6A with a Dam variant with less specific target site recognition, may be used to obtain higher resolution data.[5]
Cell-type specific methods
A major advantage of DamID over ChIP seq is that profiling of protein binding sites can be assayed in a particular cell type in vivo without requiring the physical separation of a subpopulation of cells. This allows for investigation into developmental or physiological processes in animal models.
Targeted DamID
The targeted DamID (TaDa) approach uses the phenomenon of ribosome reinitiation to express Dam-fusion proteins at appropriately low levels for DamID (i.e. Dam is non-saturating, thus avoiding toxicity). This construct can be combined with cell-type specific promoters resulting in tissue-specific methylation.[6][7] This approach can be used to assay transcription factor binding in a cell type of interest or alternatively, dam can be fused to Pol II subunits to determine binding of RNA polymerase and thus infer cell-specific gene expression. Targeted DamID has been demonstrated in Drosophila and mouse[8][9] cells.
FRT/FLP-out DamID
Cell-specific DamID can also be achieved using recombination mediated excision of a transcriptional terminator cassette upstream of the Dam-fusion protein.[10] The terminator cassette is flanked by FRT recombination sites which can be removed when combined with tissue specific expression of FLP recombinase. Upon removal of the cassette, the Dam-fusion is expressed at low levels under the control of a basal promoter.
Variants
As well as detection of standard protein-DNA interactions, DamID can be used to investigate other aspects of chromatin biology.
Split DamID
This method can be used to detect co-binding of two factors to the same genomic locus. The Dam methylase may be expressed in two halves which are fused to different proteins of interest. When both proteins bind to the same region of DNA, the Dam enzyme is reconstituted and is able to methylate the surrounding GATC sites.[11]
Chromatin accessibility
Due to the high activity of the enzyme, expression of untethered Dam results in methylation of all regions of accessible chromatin.[12][13] This approach can be used as an alternative to ATAC-seq or DNAse-seq. When combined with cell-type specific DamID methods, expression of untethered Dam can be used to identify cell-type specific promoter or enhancer regions.
RNA-DNA interactions
A DamID variant known as RNA-DamID can be used to detect interactions between RNA molecules and DNA.[14] This method relies on the expression of a Dam-MCP fusion protein which is able to bind to an RNA that has been modified with MS2 stem-loops. Binding of the Dam-fusion protein to the RNA results in detectable methylation at sites of RNA binding to the genome.
Long-range regulatory interactions
DNA sequences distal to a protein binding site may be brought into physical proximity through looping of chromosomes. For example, such interactions mediate enhancer and promoter function. These interactions can be detected through the action of Dam methylation. If Dam is targeted to a specific known DNA locus, distal sites brought into proximity due to the 3D configuration of the DNA will also be methylated and can be detected as in conventional DamID.[15]
Single cell DamID
DamID is usually performed on around 10,000 cells,[16] (although it has been demonstrated with fewer[6]). This means that the data obtained represents the average binding, or probability of a binding event across that cell population. A DamID protocol for single cells has also been developed and applied to human cells.[17] Single cell approaches can highlight the heterogeneity of chromatin associations between cells.
References
- ↑ van Steensel B, Henikoff S (April 2000). "Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase". Nature Biotechnology. 18 (4): 424–8. doi:10.1038/74487. PMID 10748524. S2CID 30350384.
- ↑ Aughey GN, Southall TD (January 2016). "Dam it's good! DamID profiling of protein-DNA interactions". Wiley Interdisciplinary Reviews: Developmental Biology. 5 (1): 25–37. doi:10.1002/wdev.205. PMC 4737221. PMID 26383089.
- ↑ Shi, Yang; He, Chuan; Aravind, L.; Hsu, Chih-Hung; Aristizábal-Corrales, David; Liu, Jianzhao; Sendinc, Erdem; Gu, Lei; Blanco, Mario Andres (2015-05-07). "DNA Methylation on N6-Adenine in C. elegans". Cell. 161 (4): 868–878. doi:10.1016/j.cell.2015.04.005. ISSN 0092-8674. PMC 4427530. PMID 25936839.
- ↑ Brooks JE, Roberts RJ (February 1982). "Modification profiles of bacterial genomes". Nucleic Acids Research. 10 (3): 913–34. doi:10.1093/nar/10.3.913. PMC 326211. PMID 6278441.
- ↑ Xiao R, Roman-Sanchez R, Moore DD (April 2010). "DamIP: a novel method to identify DNA binding sites in vivo". Nuclear Receptor Signaling. 8: e003. doi:10.1621/nrs.08003. PMC 2858267. PMID 20419059.
- 1 2 Southall TD, Gold KS, Egger B, Davidson CM, Caygill EE, Marshall OJ, Brand AH (July 2013). "Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells". Developmental Cell. 26 (1): 101–12. doi:10.1016/j.devcel.2013.05.020. PMC 3714590. PMID 23792147.
- ↑ Marshall OJ, Southall TD, Cheetham SW, Brand AH (September 2016). "Cell-type-specific profiling of protein-DNA interactions without cell isolation using targeted DamID with next-generation sequencing". Nature Protocols. 11 (9): 1586–98. doi:10.1038/nprot.2016.084. hdl:10044/1/31094. PMC 7032955. PMID 27490632.
- ↑ Tosti L, Ashmore J, Tan BS, Carbone B, Mistri TK, Wilson V, Tomlinson SR, Kaji K (April 2018). "Mapping transcription factor occupancy using minimal numbers of cells in vitro and in vivo". Genome Research. 28 (4): 592–605. doi:10.1101/gr.227124.117. PMC 5880248. PMID 29572359.
- ↑ Cheetham, Seth W.; Gruhn, Wolfram H.; van den Ameele, Jelle; Krautz, Robert; Southall, Tony D.; Kobayashi, Toshihiro; Surani, M. Azim; Brand, Andrea H. (2018-09-05). "Targeted DamID reveals differential binding of mammalian pluripotency factors". Development. 145 (20): dev.170209. doi:10.1242/dev.170209. ISSN 1477-9129. PMC 6215400. PMID 30185410.
- ↑ Pindyurin AV, Pagie L, Kozhevnikova EN, van Arensbergen J, van Steensel B (July 2016). "Inducible DamID systems for genomic mapping of chromatin proteins in Drosophila". Nucleic Acids Research. 44 (12): 5646–57. doi:10.1093/nar/gkw176. PMC 4937306. PMID 27001518.
- ↑ Hass MR, Liow HH, Chen X, Sharma A, Inoue YU, Inoue T, Reeb A, Martens A, Fulbright M, Raju S, Stevens M, Boyle S, Park JS, Weirauch MT, Brent MR, Kopan R (August 2015). "SpDamID: Marking DNA Bound by Protein Complexes Identifies Notch-Dimer Responsive Enhancers". Molecular Cell. 59 (4): 685–97. doi:10.1016/j.molcel.2015.07.008. PMC 4553207. PMID 26257285.
- ↑ Wines DR, Talbert PB, Clark DV, Henikoff S (1996). "Introduction of a DNA methyltransferase into Drosophila to probe chromatin structure in vivo". Chromosoma. 104 (5): 332–40. doi:10.1007/BF00337221. PMID 8575244. S2CID 22777948.
- ↑ Aughey GN, Estacio Gomez A, Thomson J, Yin H, Southall TD (February 2018). "CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo". eLife. 7. doi:10.7554/eLife.32341. PMC 5826290. PMID 29481322.
- ↑ Cheetham SW, Brand AH (January 2018). "RNA-DamID reveals cell-type-specific binding of roX RNAs at chromatin-entry sites". Nature Structural & Molecular Biology. 25 (1): 109–114. doi:10.1038/s41594-017-0006-4. PMC 5813796. PMID 29323275.
- ↑ Cléard F, Moshkin Y, Karch F, Maeda RK (August 2006). "Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification". Nature Genetics. 38 (8): 931–5. doi:10.1038/ng1833. PMID 16823379. S2CID 22366940.
- ↑ Marshall, Owen J.; Southall, Tony D.; Cheetham, Seth W.; Brand, Andrea H. (September 2016). "Cell-type-specific profiling of protein-DNA interactions without cell isolation using targeted DamID with next-generation sequencing". Nature Protocols. 11 (9): 1586–1598. doi:10.1038/nprot.2016.084. hdl:10044/1/31094. ISSN 1750-2799. PMC 7032955. PMID 27490632.
- ↑ Kind, Jop; Pagie, Ludo; de Vries, Sandra S.; Nahidiazar, Leila; Dey, Siddharth S.; Bienko, Magda; Zhan, Ye; Lajoie, Bryan; de Graaf, Carolyn A. (2015-09-24). "Genome-wide maps of nuclear lamina interactions in single human cells". Cell. 163 (1): 134–147. doi:10.1016/j.cell.2015.08.040. ISSN 1097-4172. PMC 4583798. PMID 26365489.
Further reading
- Vogel MJ, Peric-Hupkes D, van Steensel B (2007). "Detection of in vivo protein-DNA interactions using DamID in mammalian cells". Nature Protocols. 2 (6): 1467–78. doi:10.1038/nprot.2007.148. PMC 7032955. PMID 17545983.
- Greil F, Moorman C, van Steensel B (2006). DamID: mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Methods in Enzymology. Vol. 410. pp. 342–59. doi:10.1016/S0076-6879(06)10016-6. ISBN 9780121828158. PMID 16938559.
- Germann S, Juul-Jensen T, Letarnec B, Gaudin V (October 2006). "DamID, a new tool for studying plant chromatin profiling in vivo, and its use to identify putative LHP1 target loci". The Plant Journal. 48 (1): 153–63. doi:10.1111/j.1365-313X.2006.02859.x. PMID 16972870.