I'm using Python to simulate a process that takes place on directed graphs. I would like to produce an animation of this process.
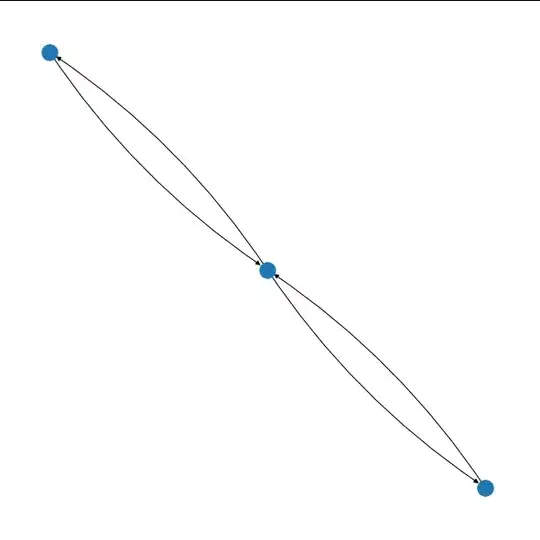
The problem that I've run into is that most Python graph visualization libraries combine pairs of directed edges into a single edge. For example, NetworkX draws only two edges when displaying the following graph, whereas I would like to display each of the four edges separately:
import networkx as nx
import matplotlib.pyplot as plt
G = nx.MultiDiGraph()
G.add_edges_from([
(1, 2),
(2, 3),
(3, 2),
(2, 1),
])
plt.figure(figsize=(8,8))
nx.draw(G)
I would like to display something like this, with each parallel edge drawn separately:
The question R reciprocal edges in igraph in R seems to deal with the same issue, but the solution there is for the R igraph library, not the Python one.
Is there an easy way to produce this style of plot using an existing Python graph visualization library? It would be a bonus if it could support multigraphs.
I'm open to solutions that invoke an external program to produce the images. I'd like to generate a whole series of animation frames, so the solution must be automated.