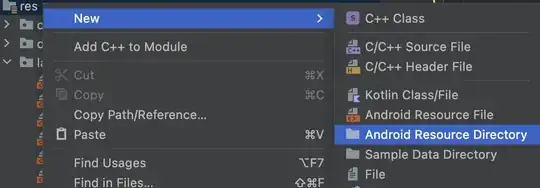
My software should judge spectrum bands, and given the location of the bands, find the peak point and width of the bands.
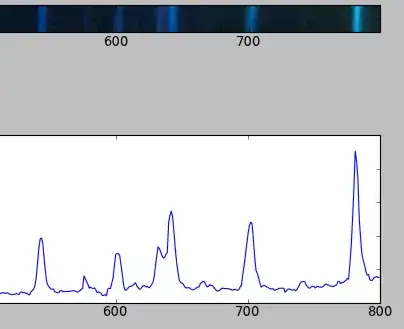
I learned to take the projection of the image and to find width of each peak.
But I need a better way to find the projection.
The method I used reduces a 1600-pixel wide image (eg 1600X40) to a 1600-long sequence. Ideally I would want to reduce the image to a 10000-long sequence using the same image.
I want a longer sequence as 1600 points provide too low resolution. A single point causes a large difference (there is a 4% difference if a band is judged from 18 to 19) to the measure.
How do I get a longer projection from the same image?
Code I used: https://stackoverflow.com/a/9771560/604511
import Image
from scipy import *
from scipy.optimize import leastsq
# Load the picture with PIL, process if needed
pic = asarray(Image.open("band2.png"))
# Average the pixel values along vertical axis
pic_avg = pic.mean(axis=2)
projection = pic_avg.sum(axis=0)
# Set the min value to zero for a nice fit
projection /= projection.mean()
projection -= projection.min()