I have a very simple question causing me to bang my head on the wall.
I would like to scale the y-axis of my histogram to reflect the proportion (0 to 1) that each bin makes up, instead of having the area of the bars sum to 1, as using y=..density.. does, or having the highest bar be 1, as y=..ncount.. does.
My input is a list of names and values, formatted like so:
name value
A 0.0000354
B 0.00768
C 0.00309
D 0.000123
One of my failed attempts:
library(ggplot2)
mydataframe < read.delim(mydata)
ggplot(mydataframe, aes(x = value)) +
geom_histogram(aes(x=value,y=..density..))
This gives me a histogram with area 1, but heights of 2000 and 1000:
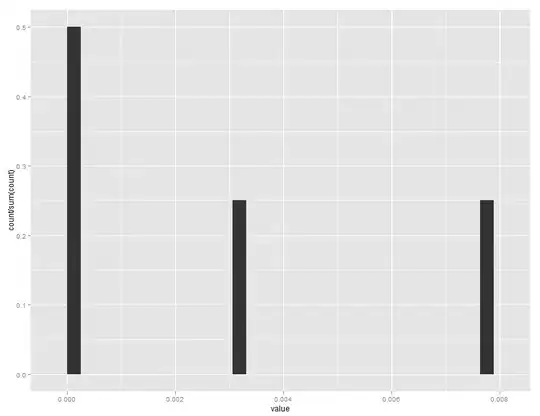
and y=..ncount.. gives me a histogram with highest bar 1.0, and rest scaled to it:
but I would like to have the first bar have a height of 0.5, and the other two 0.25.
R does not recognize these uses of scale_y_continuous either.
scale_y_continuous(formatter="percent")
scale_y_continuous(labels = percent)
scale_y_continuous(expand=c(1/(nrow(mydataframe)-1),0)
Thank you for any help.