I'm new to R and have the following challenge;
I want to create a visualization that basically combines 2 kind of 'heatmaps' in order to visualize at what times there are truly dark skies (for astronomy). For this I want to have a heatmap that visualizes the brightness of the moon based on the moonrise and moonset times and the phase of the moon. On this then we can plot a 'band'like heatmap for the time the sun is up with some transparency. I'm not sure if this is going to work visualy or if I need to find some other solution, however this seems like a good challenge to get into R some more. But I could use some pointers as I'm stuck already loading the matrix of size 24(hours) x 31(days) with all the 720 values. When trying to create a basic data.frame from the vectors I get the error that the number of rows are inconsistent.
Furthermore I have some heatmap examples working already, but I'm not sure how to combine 2 of them in the same plot like I described.
As an illustration the current 'heatmap' as it is in excel
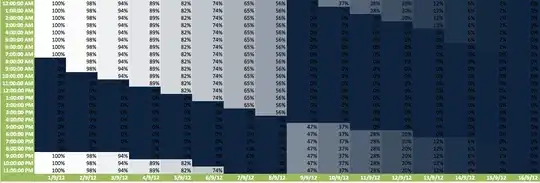
And some data:
MOON
moon <- structure(list(X1.9.12 = structure(c(2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L), .Label = c("0%", "100%"), class = "factor"), X2.9.12 = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L), .Label = c("0%", "98%"), class = "factor"),
X3.9.12 = structure(c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L
), .Label = c("0%", "94%"), class = "factor"), X4.9.12 = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L), .Label = c("0%", "89%"), class = "factor"),
X5.9.12 = structure(c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L
), .Label = c("0%", "82%"), class = "factor"), X6.9.12 = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L), .Label = c("0%", "74%"), class = "factor"),
X7.9.12 = structure(c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L
), .Label = c("0%", "65%"), class = "factor"), X8.9.12 = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), .Label = c("0%", "56%"), class = "factor"),
X9.9.12 = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L
), .Label = c("0%", "47%"), class = "factor"), X10.9.12 = structure(c(2L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("0%", "37%"), class = "factor"),
X11.9.12 = structure(c(2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L
), .Label = c("0%", "28%"), class = "factor"), X12.9.12 = structure(c(2L,
2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("0%", "20%"), class = "factor"),
X13.9.12 = structure(c(2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L
), .Label = c("0%", "12%"), class = "factor"), X14.9.12 = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L), .Label = c("0%", "6%"), class = "factor"),
X15.9.12 = structure(c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L
), .Label = c("0%", "2%"), class = "factor"), X16.9.12 = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), .Label = "0%", class = "factor"),
X17.9.12 = structure(c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L
), .Label = c("0%", "1%"), class = "factor")), .Names = c("X1.9.12",
"X2.9.12", "X3.9.12", "X4.9.12", "X5.9.12", "X6.9.12", "X7.9.12",
"X8.9.12", "X9.9.12", "X10.9.12", "X11.9.12", "X12.9.12", "X13.9.12",
"X14.9.12", "X15.9.12", "X16.9.12", "X17.9.12"), class = "data.frame", row.names = c("0:00:00",
"1:00:00", "2:00:00", "3:00:00", "4:00:00", "5:00:00", "6:00:00",
"7:00:00", "8:00:00", "9:00:00", "10:00:00", "11:00:00", "12:00:00",
"13:00:00", "14:00:00", "15:00:00", "16:00:00", "17:00:00", "18:00:00",
"19:00:00", "20:00:00", "21:00:00", "22:00:00", "23:00:00"))
SUN
September
Day Sunrise Sunset
1 6:52 20:26
2 6:54 20:24
3 6:56 20:22
4 6:57 20:20
5 6:59 20:17
6 7:00 20:15
7 7:02 20:13
8 7:04 20:10
9 7:05 20:08
10 7:07 20:06
11 7:08 20:05
12 7:09 20:02
13 7:11 20:00
14 7:13 19:58
15 7:14 19:55
16 7:16 19:53
17 7:17 19:51
18 7:19 19:48
19 7:21 19:46
20 7:22 19:44
21 7:25 19:40
22 7:26 19:38
23 7:28 19:35
24 7:30 19:33
25 7:31 19:31
26 7:33 19:28
27 7:35 19:26
28 7:36 19:24
29 7:38 19:21
30 7:40 19:19