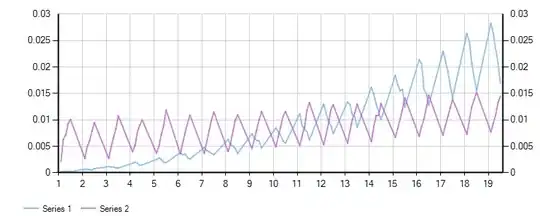
I'm trying to do something similar to what's outlined in this post: MATLAB, Filling in the area between two sets of data, lines in one figure but running into a roadblock. I'm trying to shade the area of a graph that represents the mean +/- standard deviation. The variable definitions are a bit complicated but it boils down to this code, and when plotted without shading, I get the screenshot below:
x = linspace(0, 100, 101)';
mean = torqueRnormMean(:,1);
meanPlusSTD = torqueRnormMean(:,1) + torqueRnormStd(:,1);
meanMinusSTD = torqueRnormMean(:,1) - torqueRnormStd(:,1);
plot(x, mean, 'k', 'LineWidth', 2)
plot(x, meanPlusSTD, 'k--')
plot(x, meanMinusSTD, 'k--')
But when I try to implement shading just on the lower half of the graph (between mean and meanMinusSTD) by adding the code below, I get a plot that looks like this:
fill( [x fliplr(x)], [mean fliplr(meanMinusSTD)], 'y', 'LineStyle','--');
It's obviously not shading the correct area of the graph, and new near-horizontal lines are being created close to 0 that are messing with the shading.
Any thoughts? I'm stumped.