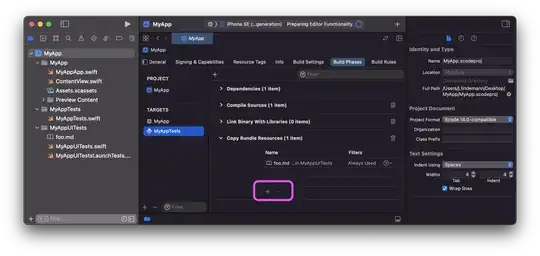
2nd Extension to BenVoigt's answer
My suggestion for one kernel filter is with convolution of relative random error
data(find(data ~= 0)) = sin(pi .* data(find(data ~= 0))) ./ (pi*data(find(data ~= 0)));
data(find(data == 0)) = 1; % removing lastly the discontinuity
data1 = data + 0.0000001 * mean(abs(data(:))) * randn(size(data));
data = conv(data, data1);
Is this what BenVoigt means by the kernel filter for distribution?
This gives results like
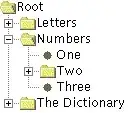
where the resolution is still a problem.
The central peaks tend to multiply easily if I resize the window.
I had old code active in the above picture but it did not change the result.
The above code is still not enough for the kernel filter of the display.
Probably, some filter functions has to be applied to the time and frequency axis separately still, something like:
F1 = filter2(B,T); % you need a different kernel filter because resolution is lower
T = filter2(B,F);
F = F1;
These filters mess up the values on the both axis.
I need to understand them better to fix this.
But first to understand if they are the right way to go.
The figure has be resized still.
The size of the data was 5001x1 double and those of F and T 13635x1 double.
So I think I should resize lastly after setting axis, labels and title by
imresize(image, [13635 13635], 'bilinear');
since the distirbution is bilinear.