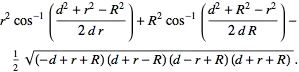This a rather peculiar question. Normally probably it's faster to "photoshop" one than rastering the plots into images etc (I meant, measured by how much human effort it takes).
But since your question has a fun component in it, below I provide one shoddy solution.
The starting point is the example you provided. I use some R code to extract the two faces, and then combine them with two plots (any plots). The plots will be saved to PNG, and then loaded into R as a rastered image.
Eventually you combine the four facets and make one final plot.
library(png)
library(ggplot2)
# load PNG file, and reduce dimension to 1.
# there's no sanity check to verify how many channels a PNG file has.
# potentially there can be 1 (grayscale), 3 (RGB) and 4 (RGBA).
# Here I assume that PNG file f has 4 channels.
load_png <- function(f) {
d <- readPNG(f)
# CCIR 601
rgb.weights <- c(0.2989, 0.5870, 0.1140)
grayscale <- matrix(apply(d[,,-4], -3,
function(rgb) rgb %*% rgb.weights),
ncol=dim(d)[2], byrow=T)
grayscale
}
# the image you provided as an example,
# used to extract the two emoicons
img <- load_png("3anUH.png")
# convert a grayscale matrix into a data.frame,
# facilitating plotting by ggplot
melt_grayscale <- function(d) {
w <- ncol(d)
h <- nrow(d)
coords <- expand.grid(1:w, 1:h)
df <- cbind(coords, as.vector(d))
names(df) <- c("x", "y", "gs")
# so that smallest Y is at the top
df$y <- h - df$y + 1
df
}
plot_grayscale <- function(d, melt=F) {
df <- melt_grayscale(d)
ggplot(df) + geom_raster(aes(x=x, y=y, fill=gs)) + scale_fill_continuous(low="#000000", high="#ffffff")
}
ggplot_blank <- function(x, y) {
# to plot a graph without any axis, grid and legend
# otherwise it would look weird when performing facet_wrap()
qplot(x, y + rnorm(10), size=15) +
theme(axis.line=element_blank(),
axis.text.x=element_blank(),
axis.text.y=element_blank(),
axis.ticks=element_blank(),
axis.title.x=element_blank(),
axis.title.y=element_blank(),
legend.position="none",
panel.background=element_blank(),
panel.border=element_blank(),
panel.grid.major=element_blank(),
panel.grid.minor=element_blank(),
plot.background=element_blank())
}
# extract the two faces
#offset <- c(50, 40)
img0 <- img[50:200, 40:190]
img1 <- img[210:360, 40:190]
plot_grayscale(img0)
plot_grayscale(img1)
At this point we have img0 that looks like: 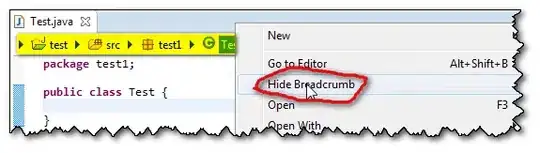 .
.
You can probably adjust the subsetting offset to getting a cleaner cut.
Next we want to draw two plots, and save the plot to PNGs which can be loaded later.
# now plot two PNGs using ggplot
png(file="p0.png", width=150, height=150)
ggplot_blank(1:10, 10:1 + rnorm(10))
dev.off()
png(file="p1.png", width=150, height=150)
ggplot_blank(1:10, rnorm(10, 10))
dev.off()
p0 <- load_png("p0.png")
p1 <- load_png("p1.png")
# combine PNG grayscale matrices together
# into a melted data.frame, but with an extra column to
# identify which panel does this pixel belong to.
combine.plots <- function(l) {
panel <- 0
do.call(rbind, lapply(l, function(m){
panel <<- panel + 1
cbind(melt_grayscale(m), panel)
}))
}
plots <- combine.plots(list(img0, img1, p0, p1))
ggplot(plots) + geom_raster(aes(x=x, y=y, fill=gs)) +
scale_fill_continuous(low="#000000", high="#ffffff") + facet_wrap(~panel)
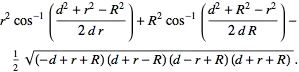
Of course the obvious drawback is that you only have grayscale image with above example.
It would be much trickier if you want RGB color in your final plot.
You can probably do what GIF format is doing: indexed color.
Basically you:
- Discretize your RGB values into 256 or 512 colors
- Create a vector of the discretized colors
- Replaces
scale_fill_continous with scale_fill_manual and let values equal to the color vector you created above.
EDIT: Combine geom_raster with geom_point.
Above solution used png() function to first save the plot to a PNG file (involving rasterization), and then loaded the rasterized images into R. This process could potentially lead to resolution loss, if, say, the two images shown in the top panels are themselves of low resolution.
We can modify above solution to combine geom_point with geom_raster, where the former is used for rendering plots and the latter for rendering images.
The only issue here, (assuming that the two images to display have the same resolution, denoted by w x h, where w is the width and h is the height), is that facet_wrap will enforce all panels to have the same X/Y-limits.
So, we need to rescale the plots to the same limits (w x h) before plotting them.
Below is the modified R code for combining plots and images:
library(png)
library(ggplot2)
load_png <- function(f) {
d <- readPNG(f)
# CCIR 601
rgb.weights <- c(0.2989, 0.5870, 0.1140)
grayscale <- matrix(apply(d[,,-4], -3,
function(rgb) rgb %*% rgb.weights),
ncol=dim(d)[2], byrow=T)
grayscale
}
# the image you provided as an example,
# used to extract the two emoicons
img <- load_png("3anUH.png")
# convert a grayscale matrix into a data.frame,
# facilitating plotting by ggplot
melt_grayscale <- function(d) {
w <- ncol(d)
h <- nrow(d)
coords <- expand.grid(1:w, 1:h)
df <- cbind(coords, as.vector(d))
names(df) <- c("x", "y", "gs")
df$y <- h - df$y + 1
df
}
plot_grayscale <- function(d, melt=F) {
df <- melt_grayscale(d)
ggplot(df) + geom_raster(aes(x=x, y=y, fill=gs)) + scale_fill_continuous(low="#000000", high="#ffffff")
}
ggplot_blank <- function(x, y) {
# to plot a graph without any axis, grid and legend
qplot(x, y + rnorm(10), size=15) +
theme(axis.line=element_blank(),
axis.text.x=element_blank(),
axis.text.y=element_blank(),
axis.ticks=element_blank(),
axis.title.x=element_blank(),
axis.title.y=element_blank(),
legend.position="none",
panel.background=element_blank(),
panel.border=element_blank(),
panel.grid.major=element_blank(),
panel.grid.minor=element_blank(),
plot.background=element_blank())
}
# extract the two faces
#offset <- c(50, 40)
img0 <- img[50:200, 40:190]
img1 <- img[210:360, 40:190]
plot_grayscale(img0)
plot_grayscale(img1)
# now plot two PNGs using ggplot
png(file="p0.png", width=300, height=300)
ggplot_blank(1:10, 10:1 + rnorm(10))
dev.off()
png(file="p1.png", width=300, height=300)
ggplot_blank(1:10, rnorm(10, 10))
dev.off()
p0 <- load_png("p0.png")
p1 <- load_png("p1.png")
combine.plots <- function(l) {
panel <- 0
do.call(rbind, lapply(l, function(m){
panel <<- panel + 1
cbind(melt_grayscale(m), panel)
}))
}
rescale.plots <- function(x, y, w, h, panel) {
# need to rescale plots to the same scale (w x h)
x <- (x - min(x)) / (max(x) - min(x)) * w
y <- (y - min(y)) / (max(y) - min(y)) * h
data.frame(x=x, y=y, panel=panel)
}
imgs <- combine.plots(list(img0, img1))
# combine two plots, with proper rescaling
plots <- rbind(
rescale.plots(1:100, 100:1 + rnorm(100), 150, 150, panel=3),
rescale.plots(1:100, rnorm(100), 150, 150, panel=4)
)
ggplot() + geom_raster(data=imgs, aes(x=x, y=y, fill=gs)) + geom_point(data=plots, aes(x=x, y=y)) +
facet_wrap(~panel) + scale_fill_continuous(low="#000000", high="#ffffff")
which will give you:
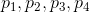
You may not visually detect the immediate difference, but it will be obvious when you resize the output image in R.

 .
.