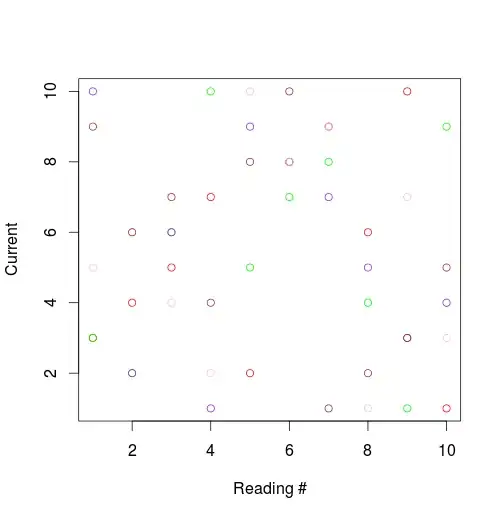
I'm plotting multiple data series.
colos=c('red','green','purple','pink','brown')
par(new=F)
for (i in 1:5)
{
plot(dat[[i+1]],col=colos[i],cex=marksize,xlab='Reading #',ylab = 'Current')
par(new=T)
}
My plot looks like this:
Is there a way I can overwrite the plot axis with each iteration, but not overwrite the plotted points?