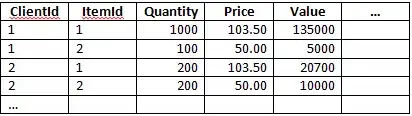
My data is:
>>> ts = pd.TimeSeries(data,indexconv)
>>> tsgroup = ts.resample('t',how='sum')
>>> tsgroup
2014-11-08 10:30:00 3
2014-11-08 10:31:00 4
2014-11-08 10:32:00 7
[snip]
2014-11-08 10:54:00 5
2014-11-08 10:55:00 2
Freq: T, dtype: int64
>>> tsgroup.plot()
>>> plt.show()
indexconv are strings converted using datetime.strptime.
The plot is very edgy like this (these aren't my actual plots):

How can I smooth it out like this:

I know about scipy.interpolate mentioned in this article (which is where I got the images from), but how can I apply it for Pandas time series?
I found this great library called Vincent that deals with Pandas, but it doesn't support Python 2.6.