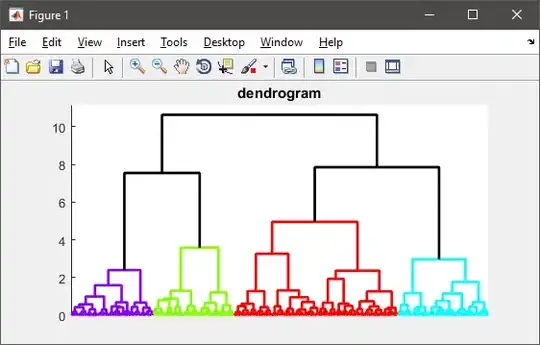
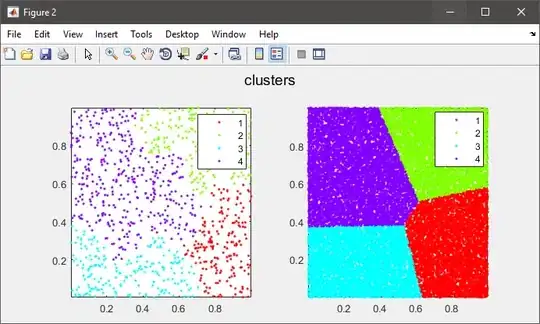
As mentioned by others, hierarchical clustering needs to calculate the pairwise distance matrix which is too big to fit in memory in your case.
Try using the K-Means algorithm instead:
numClusters = 4;
T = kmeans(X, numClusters);
Alternatively you can select a random subset of your data and use as input to the clustering algorithm. Next you compute the cluster centers as mean/median of each cluster group. Finally for each instance that was not selected in the subset, you simply compute its distance to each of the centroids and assign it to the closest one.
Here's a sample code to illustrate the idea above:
%# random data
X = rand(25000, 2);
%# pick a subset
SUBSET_SIZE = 1000; %# subset size
ind = randperm(size(X,1));
data = X(ind(1:SUBSET_SIZE), :);
%# cluster the subset data
D = pdist(data, 'euclid');
T = linkage(D, 'ward');
CUTOFF = 0.6*max(T(:,3)); %# CUTOFF = 5;
C = cluster(T, 'criterion','distance', 'cutoff',CUTOFF);
K = length( unique(C) ); %# number of clusters found
%# visualize the hierarchy of clusters
figure(1)
h = dendrogram(T, 0, 'colorthreshold',CUTOFF);
set(h, 'LineWidth',2)
set(gca, 'XTickLabel',[], 'XTick',[])
%# plot the subset data colored by clusters
figure(2)
subplot(121), gscatter(data(:,1), data(:,2), C), axis tight
%# compute cluster centers
centers = zeros(K, size(data,2));
for i=1:size(data,2)
centers(:,i) = accumarray(C, data(:,i), [], @mean);
end
%# calculate distance of each instance to all cluster centers
D = zeros(size(X,1), K);
for k=1:K
D(:,k) = sum( bsxfun(@minus, X, centers(k,:)).^2, 2);
end
%# assign each instance to the closest cluster
[~,clustIDX] = min(D, [], 2);
%#clustIDX( ind(1:SUBSET_SIZE) ) = C;
%# plot the entire data colored by clusters
subplot(122), gscatter(X(:,1), X(:,2), clustIDX), axis tight