For a "write this code for me" question showing no effort, you certainly have a lot of specific demands. This doesn't fit your criteria, but maybe someone will find it useful in base graphics
The plot in the center panel can be just about anything so long as there is one plot per line and kindasorta fits within each. (Actually that's not true, any kind of plot can go in that panel if you want since it's just a normal plotting window). There are three examples in this code: points, box plots, lines.

This is the input data. Just a generic list and indices for "headers" so much better IMO than "directly using a regression object."
## indices of headers
idx <- c(1,5,7,22)
l <- list('Make/model' = rownames(mtcars),
'No. of\ncycles' = mtcars$cyl,
MPG = mtcars$mpg)
l[] <- lapply(seq_along(l), function(x)
ifelse(seq_along(l[[x]]) %in% idx, l[[x]], paste0(' ', l[[x]])))
# List of 3
# $ Make/model : chr [1:32] "Mazda RX4" " Mazda RX4 Wag" " Datsun 710" " Hornet 4 Drive" ...
# $ No. of
# cycles: chr [1:32] "6" " 6" " 4" " 6" ...
# $ MPG : chr [1:32] "21" " 21" " 22.8" " 21.4" ...
I realize this code generates a pdf. I didn't feel like changing it to an image to upload, so I converted it with imagemagick
## choose the type of plot you want
pl <- c('point','box','line')[1]
## extra (or less) c(bottom, left, top, right) spacing for additions in margins
pad <- c(0,0,0,0)
## default padding
oma <- c(1,1,2,1)
## proportional size of c(left, middle, right) panels
xfig = c(.25,.45,.3)
## proportional size of c(caption, main plot)
yfig = c(.15, .85)
cairo_pdf('~/desktop/pl.pdf', height = 9, width = 8)
x <- l[-3]
lx <- seq_along(x[[1]])
nx <- length(lx)
xcf <- cumsum(xfig)[-length(xfig)]
ycf <- cumsum(yfig)[-length(yfig)]
plot.new()
par(oma = oma, mar = c(0,0,0,0), family = 'serif')
plot.window(range(seq_along(x)), range(lx))
## bars -- see helper fn below
par(fig = c(0,1,ycf,1), oma = par('oma') + pad)
bars(lx)
## caption
par(fig = c(0,1,0,ycf), mar = c(0,0,3,0), oma = oma + pad)
p <- par('usr')
box('plot')
rect(p[1], p[3], p[2], p[4], col = adjustcolor('cornsilk', .5))
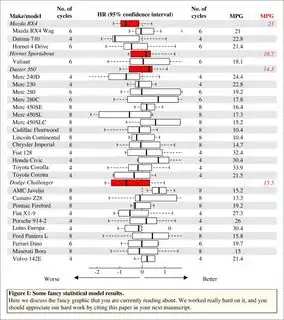
mtext('\tFigure I: Some fancy statistical model results.',
adj = 0, font = 2, line = -1)
mtext(paste('\tHere we discuss the fancy graphic that you are currently reading',
'about. We worked really hard on it, and you\n\tshould appreciate',
'our hard work by citing this paper in your next manuscript.'),
adj = 0, line = -3)
## left panel -- select two columns
lp <- l[1:2]
par(fig = c(0,xcf[1],ycf,1), oma = oma + vec(pad, 0, 4))
plot_text(lp, c(1,2),
adj = rep(0:1, c(nx, nx)),
font = vec(1, 3, idx, nx),
col = c(rep(1, nx), vec(1, 'transparent', idx, nx))
) -> at
vtext(unique(at$x), max(at$y) + c(1,1.5), names(lp),
font = 2, xpd = NA, adj = c(0,1))
## right panel -- select three columns
rp <- l[c(2:3,3)]
par(fig = c(tail(xcf, -1),1,ycf,1), oma = oma + vec(pad, 0, 2))
plot_text(rp, c(1,2),
col = c(rep(vec(1, 'transparent', idx, nx), 2),
vec('transparent', 2, idx, nx)),
font = vec(1, 3, idx, nx),
adj = rep(c(NA,NA,1), each = nx)
) -> at
vtext(unique(at$x), max(at$y) + c(1.5,1,1), names(rp),
font = 2, xpd = NA, adj = c(NA, NA, 1), col = c(1,1,2))
## middle panel -- some generic plot
par(new = TRUE, fig = c(xcf[1], xcf[2], ycf, 1),
mar = c(0,2,0,2), oma = oma + vec(pad, 0, c(2,4)))
set.seed(1)
xx <- rev(rnorm(length(lx)))
yy <- rev(lx)
plot(xx, yy, ann = FALSE, axes = FALSE, type = 'n',
panel.first = {
segments(0, 0, 0, nx, lty = 'dashed')
},
panel.last = {
## option 1: points, confidence intervals
if (pl == 'point') {
points(xx, yy, pch = 15, col = vec(1, 2, idx, nx))
segments(xx * .5, yy, xx * 1.5, yy, col = vec(1, 2, idx, nx))
}
## option 2: boxplot, distributions
if (pl == 'box')
boxplot(rnorm(200) ~ rep_len(1:nx, 200), at = nx:1,
col = vec(par('bg'), 2, idx, nx),
horizontal = TRUE, axes = FALSE, add = TRUE)
## option 3: trend lines
if (pl == 'line') {
for (ii in 1:nx) {
n <- sample(40, 1)
wh <- which(nx:1 %in% ii)
lines(cumsum(rep(.1, n)) - 2, wh + cumsum(runif(n, -.2, .2)), xpd = NA,
col = (ii %in% idx) + 1L, lwd = c(1,3)[(ii %in% idx) + 1L])
}
}
## final touches
mtext('HR (95% confidence interval)', font = 2, line = -.5)
axis(1, at = -3:2, tcl = 0.2, mgp = c(0,0,0))
mtext(c('Worse','Better'), side = 1, line = 1, at = c(-4, 3))
try(silent = TRUE, {
## can just replace this with graphics::arrows with minor changes
## i just like the filled ones
rawr::arrows2(-.1, -1.5, -3, size = .5, width = .5)
rawr::arrows2(0.1, -1.5, 2, size = .5, width = .5)
})
}
)
box('outer')
dev.off()
Using these four helper functions (see example use in the body)
vec <- function(default, replacement, idx, n) {
# vec(1, 0, 2:3, 5); vec(1:5, 0, 2:3)
out <- if (missing(n))
default else rep(default, n)
out[idx] <- replacement
out
}
bars <- function(x, cols = c(NA, grey(.9)), horiz = TRUE) {
# plot(1:10, type = 'n'); bars(1:10)
p <- par('usr')
cols <- vec(cols[1], cols[2], which(!x %% 2), length(x))
x <- rev(x) + 0.5
if (horiz)
rect(p[1], x - 1L, p[2], x, border = NA, col = rev(cols), xpd = NA) else
rect(x - 1L, p[3], x, p[4], border = NA, col = rev(cols), xpd = NA)
invisible()
}
vtext <- function(...) {Vectorize(text.default)(...); invisible()}
plot_text <- function(x, width = range(seq_along(x)), ...) {
# plot(col(mtcars), row(mtcars), type = 'n'); plot_text(mtcars)
lx <- lengths(x)[1]
rn <- range(seq_along(x))
sx <- (seq_along(x) - 1) / diff(rn) * diff(width) + width[1]
xx <- rep(sx, each = lx)
yy <- rep(rev(seq.int(lx)), length(x))
vtext(xx, yy, unlist(x), ..., xpd = NA)
invisible(list(x = sx, y = rev(seq.int(lx))))
}