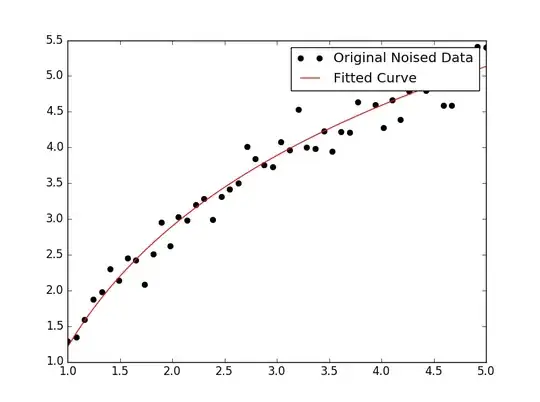
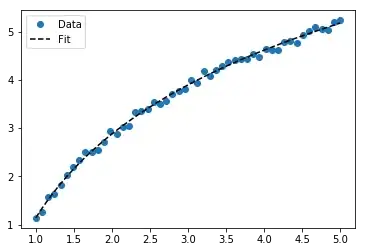
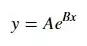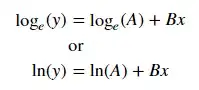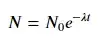For fitting y = A + B log x, just fit y against (log x).
>>> x = numpy.array([1, 7, 20, 50, 79])
>>> y = numpy.array([10, 19, 30, 35, 51])
>>> numpy.polyfit(numpy.log(x), y, 1)
array([ 8.46295607, 6.61867463])
# y ≈ 8.46 log(x) + 6.62
For fitting y = AeBx, take the logarithm of both side gives log y = log A + Bx. So fit (log y) against x.
Note that fitting (log y) as if it is linear will emphasize small values of y, causing large deviation for large y. This is because polyfit (linear regression) works by minimizing ∑i (ΔY)2 = ∑i (Yi − Ŷi)2. When Yi = log yi, the residues ΔYi = Δ(log yi) ≈ Δyi / |yi|. So even if polyfit makes a very bad decision for large y, the "divide-by-|y|" factor will compensate for it, causing polyfit favors small values.
This could be alleviated by giving each entry a "weight" proportional to y. polyfit supports weighted-least-squares via the w keyword argument.
>>> x = numpy.array([10, 19, 30, 35, 51])
>>> y = numpy.array([1, 7, 20, 50, 79])
>>> numpy.polyfit(x, numpy.log(y), 1)
array([ 0.10502711, -0.40116352])
# y ≈ exp(-0.401) * exp(0.105 * x) = 0.670 * exp(0.105 * x)
# (^ biased towards small values)
>>> numpy.polyfit(x, numpy.log(y), 1, w=numpy.sqrt(y))
array([ 0.06009446, 1.41648096])
# y ≈ exp(1.42) * exp(0.0601 * x) = 4.12 * exp(0.0601 * x)
# (^ not so biased)
Note that Excel, LibreOffice and most scientific calculators typically use the unweighted (biased) formula for the exponential regression / trend lines. If you want your results to be compatible with these platforms, do not include the weights even if it provides better results.
Now, if you can use scipy, you could use scipy.optimize.curve_fit to fit any model without transformations.
For y = A + B log x the result is the same as the transformation method:
>>> x = numpy.array([1, 7, 20, 50, 79])
>>> y = numpy.array([10, 19, 30, 35, 51])
>>> scipy.optimize.curve_fit(lambda t,a,b: a+b*numpy.log(t), x, y)
(array([ 6.61867467, 8.46295606]),
array([[ 28.15948002, -7.89609542],
[ -7.89609542, 2.9857172 ]]))
# y ≈ 6.62 + 8.46 log(x)
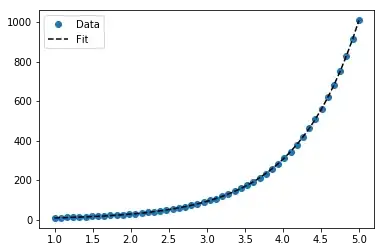
For y = AeBx, however, we can get a better fit since it computes Δ(log y) directly. But we need to provide an initialize guess so curve_fit can reach the desired local minimum.
>>> x = numpy.array([10, 19, 30, 35, 51])
>>> y = numpy.array([1, 7, 20, 50, 79])
>>> scipy.optimize.curve_fit(lambda t,a,b: a*numpy.exp(b*t), x, y)
(array([ 5.60728326e-21, 9.99993501e-01]),
array([[ 4.14809412e-27, -1.45078961e-08],
[ -1.45078961e-08, 5.07411462e+10]]))
# oops, definitely wrong.
>>> scipy.optimize.curve_fit(lambda t,a,b: a*numpy.exp(b*t), x, y, p0=(4, 0.1))
(array([ 4.88003249, 0.05531256]),
array([[ 1.01261314e+01, -4.31940132e-02],
[ -4.31940132e-02, 1.91188656e-04]]))
# y ≈ 4.88 exp(0.0553 x). much better.