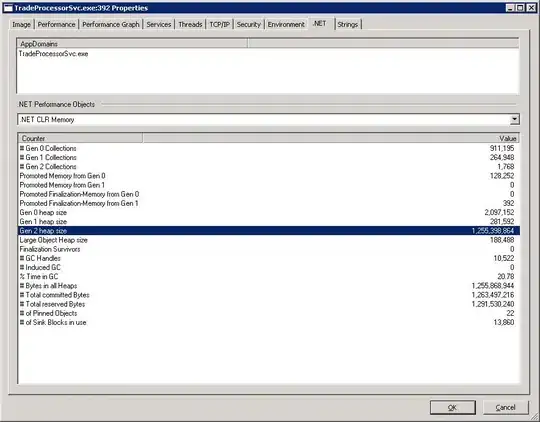
I am using RStudio to draw the histogram of the values in this data.
data = read.table("C:\\Test\\test.csv", header=TRUE, sep=",")
hist(data$a, breaks=100)
hist(data$b, breaks=100)
and got the following histograms:
1- have the y axis logged so that instead of values 0, 4000, 8000 and 12000 I'll have 0, 10, 100, 1000, 10000 and so on (log2 i.e., 0, 1, 2, 4, 8, 16, ... is also useful).
2- Have the two diagrams in a single diagram (preferably with bars of two different colors/patterns). In the resulting diagram, the two bars for each x-value would be beside each other like this:
I tried this solution but got the following error:
NULL
Warning message: In (function () : Only one RStudio graphics
device is permitted