I took both inputs, as well as my original approach and compared the results. As @eat correctly points out, the results depend on the nature of your input data. Surprisingly, concatenate beats view processing in a few instances. Each method has a sweet-spot. Here is my benchmark code:
import numpy as np
from itertools import product
def segment_and_concatenate(M, fun=None, blk_size=(16,16), overlap=(0,0)):
# truncate M to a multiple of blk_size
M = M[:M.shape[0]-M.shape[0]%blk_size[0],
:M.shape[1]-M.shape[1]%blk_size[1]]
rows = []
for i in range(0, M.shape[0], blk_size[0]):
cols = []
for j in range(0, M.shape[1], blk_size[1]):
max_ndx = (min(i+blk_size[0], M.shape[0]),
min(j+blk_size[1], M.shape[1]))
cols.append(fun(M[i:max_ndx[0], j:max_ndx[1]]))
rows.append(np.concatenate(cols, axis=1))
return np.concatenate(rows, axis=0)
from numpy.lib.stride_tricks import as_strided
def block_view(A, block= (3, 3)):
"""Provide a 2D block view to 2D array. No error checking made.
Therefore meaningful (as implemented) only for blocks strictly
compatible with the shape of A."""
# simple shape and strides computations may seem at first strange
# unless one is able to recognize the 'tuple additions' involved ;-)
shape= (A.shape[0]/ block[0], A.shape[1]/ block[1])+ block
strides= (block[0]* A.strides[0], block[1]* A.strides[1])+ A.strides
return as_strided(A, shape= shape, strides= strides)
def segmented_stride(M, fun, blk_size=(3,3), overlap=(0,0)):
# This is some complex function of blk_size and M.shape
stride = blk_size
output = np.zeros(M.shape)
B = block_view(M, block=blk_size)
O = block_view(output, block=blk_size)
for b,o in zip(B, O):
o[:,:] = fun(b);
return output
def view_process(M, fun=None, blk_size=(16,16), overlap=None):
# truncate M to a multiple of blk_size
from itertools import product
output = np.zeros(M.shape)
dz = np.asarray(blk_size)
shape = M.shape - (np.mod(np.asarray(M.shape),
blk_size))
for indices in product(*[range(0, stop, step)
for stop,step in zip(shape, blk_size)]):
# Don't overrun the end of the array.
#max_ndx = np.min((np.asarray(indices) + dz, M.shape), axis=0)
#slices = [slice(s, s + f, None) for s,f in zip(indices, dz)]
output[indices[0]:indices[0]+dz[0],
indices[1]:indices[1]+dz[1]][:,:] = fun(M[indices[0]:indices[0]+dz[0],
indices[1]:indices[1]+dz[1]])
return output
if __name__ == "__main__":
R = np.random.rand(128,128)
squareit = lambda(x):x*2
from timeit import timeit
t ={}
kn = np.array(list(product((8,16,64,128),
(128, 512, 2048, 4096)) ) )
methods = ("segment_and_concatenate",
"view_process",
"segmented_stride")
t = np.zeros((kn.shape[0], len(methods)))
for i, (k, N) in enumerate(kn):
for j, method in enumerate(methods):
t[i,j] = timeit("""Rprime = %s(R, blk_size=(%d,%d),
overlap = (0,0),
fun = squareit)""" % (method, k, k),
setup="""
from segmented_processing import %s
import numpy as np
R = np.random.rand(%d,%d)
squareit = lambda(x):x**2""" % (method, N, N),
number=5
)
print "k =", k, "N =", N #, "time:", t[i]
print (" Speed up (view vs. concat, stride vs. concat): %0.4f, %0.4f" % (
t[i][0]/t[i][1],
t[i][0]/t[i][2]))
And here are the results:
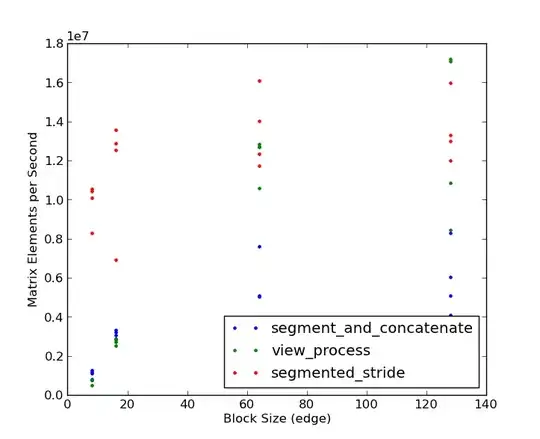 Note that the segmented stride method wins by 3-4x for small block sizes. Only at large block sizes (128 x 128) and very large matrices (2048 x 2048 and larger) does the view processing approach win, and then only by a small percentage. Based on the bake-off, it looks like @eat gets the check-mark! Thanks to both of you for good examples!
Note that the segmented stride method wins by 3-4x for small block sizes. Only at large block sizes (128 x 128) and very large matrices (2048 x 2048 and larger) does the view processing approach win, and then only by a small percentage. Based on the bake-off, it looks like @eat gets the check-mark! Thanks to both of you for good examples!
 Note that the segmented stride method wins by 3-4x for small block sizes. Only at large block sizes (128 x 128) and very large matrices (2048 x 2048 and larger) does the view processing approach win, and then only by a small percentage. Based on the bake-off, it looks like @eat gets the check-mark! Thanks to both of you for good examples!
Note that the segmented stride method wins by 3-4x for small block sizes. Only at large block sizes (128 x 128) and very large matrices (2048 x 2048 and larger) does the view processing approach win, and then only by a small percentage. Based on the bake-off, it looks like @eat gets the check-mark! Thanks to both of you for good examples!