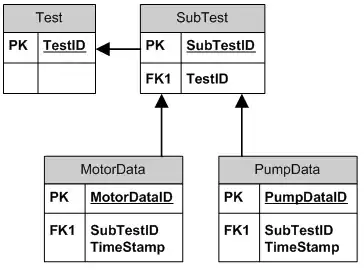
Explanation
I couldn't see your data anywhere here, but I will use the iris dataset to demonstrate how this can be achieved. I'll stick to only two examples because this takes some time to code, but once you see two examples I think it will become fairly quick to recognize what is going on. I will supply a reference at the end that will be helpful too.
Fitting Model Data
First we can fit a regression using the iris data, then turn the data into a tibble with model data using both fortify and as_tibble. I have added an index column for one of the plots later.
#### Load Library ####
library(tidyverse)
#### Fit Model ####
fit <- lm(Petal.Width ~ Petal.Length,
data = iris)
#### Turn Model into Data Frame ####
fit.data <- fortify(fit) %>%
as_tibble() %>%
mutate(.index = 1:150)
fit.data
Which gives you this:
# A tibble: 150 × 9
Petal…¹ Petal…² .hat .sigma .cooksd .fitted .resid .stdr…³ .index
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
1 0.2 1.4 0.0186 0.207 8.18e-5 0.219 -0.0190 -0.0928 1
2 0.2 1.4 0.0186 0.207 8.18e-5 0.219 -0.0190 -0.0928 2
3 0.2 1.3 0.0197 0.207 1.23e-4 0.177 0.0226 0.111 3
4 0.2 1.5 0.0176 0.207 7.86e-4 0.261 -0.0606 -0.296 4
5 0.2 1.4 0.0186 0.207 8.18e-5 0.219 -0.0190 -0.0928 5
6 0.4 1.7 0.0158 0.207 6.06e-4 0.344 0.0563 0.275 6
7 0.3 1.4 0.0186 0.207 1.49e-3 0.219 0.0810 0.396 7
8 0.2 1.5 0.0176 0.207 7.86e-4 0.261 -0.0606 -0.296 8
9 0.2 1.4 0.0186 0.207 8.18e-5 0.219 -0.0190 -0.0928 9
10 0.1 1.5 0.0176 0.207 5.53e-3 0.261 -0.161 -0.785 10
# … with 140 more rows, and abbreviated variable names ¹Petal.Width,
# ²Petal.Length, ³.stdresid
# ℹ Use `print(n = ...)` to see more rows
You can see here it gives you a lot of valuable information...residuals, fitted residuals, Cook's distance, etc. This makes it easy to plot them in ggplot2.
Plotting
The first example will be a Cook's distance plot. This takes the index of the data point and plots the columns representing their respective distance using the geom_col function. The key ingredient here is the geom_text portion. Simply subset the data and nudge it a little so it doesnt totally overlap and you can essentially label whatever you want:
#### Cooks Distance ####
fit.data %>%
ggplot(aes(x=.index,
y=.cooksd,
label=.index))+
geom_col()+
labs(x="Index",
y="Cook's Distance",
title = "Cook's Distance")+
geom_text(data=subset(fit.data,
.cooksd > .05),
nudge_y = .003)
Giving you this plot:

Another example using a similar method below plots fitted values versus their respective residuals, with an arbitrary label placed here was well:
#### Fitted vs Residuals ####
ggplot(fit.data,
aes(.fitted,
round(.resid,2),
label=round(.resid,2))) +
geom_point() +
geom_hline(yintercept = 0) +
geom_smooth(se = FALSE)+
labs(x="Fitted",
y="Residual",
title = "Fitted vs Residuals")+
geom_text(data=subset(fit.data,
.resid > .5 | .resid < -.5),
nudge_x = .09)
A slew of other examples of how to do this can be seen at this link. The customization will be up to you, but it should give you a fair idea of how to hand tailor some of these base R plots you are getting.