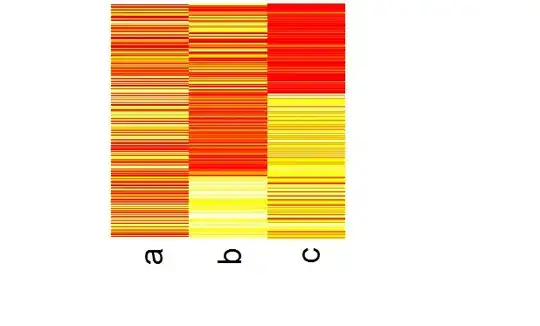
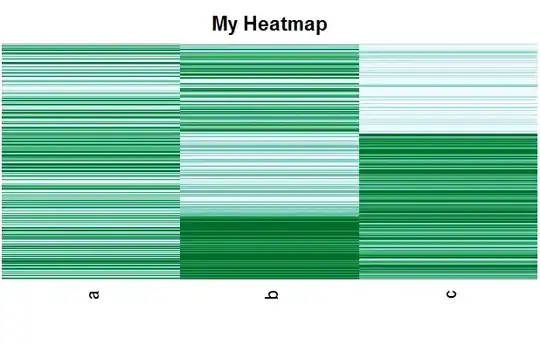
I searched a lot of questions about heatmap throughout the site and packages, but I still have a problem.
I have clustered data (kmeans/EM/DBscan..), and I want to create a heatmap by grouping the same cluster. I want the similar color patterns to be grouped in the heatmap, so generally, it looks like a block-diagonal.
I tried to order the data by the cluster number and display it,
k = kmeans(data, 3)
d = data.frame(data)
d = data.frame(d, k$cluster)
d = d[order(d$k.cluster),]
heatmap(as.matrix(d))
But, I want it to be sorted by its cluster number and looked like this:
Can I do this in R?
I searched lots of packages and tried many ways, but I still have a problem.
Thanks a lot.