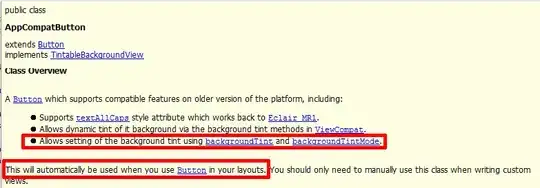
I have constructed kd-trees for two sets of points, in order to find the closest bichromatic pairing between the two sets:
The kd-trees are stored as python dictionaries, which can be found in the code below, and are passed to a function ('closest') that is meant to simultaneously recursively analyse both trees to find the closest approach between the sets. This is to prevent having to brute force the problem.
My first attempt is based on the answer to this question. With this attempt, I can't find a condition that forces the function to 'bounce back' when it hits a leaf i.e the if statement designed to return the minimum distances between the leaves and the existing minimum is never reached.
First attempt - full code provided for context, this question only pertains to the function 'closest':
from operator import itemgetter
import math
import time
import pprint
import numpy as np
# builds the trees
def build_kd_tree(ar, depth=0, k=2):
if len(ar) <= 0:
return None
axis = depth % k
sorted_ar = sorted(ar, key=itemgetter(axis))
idx = int(math.floor(len(ar)/2))
return {
'point': sorted_ar[idx],
'left': build_kd_tree(sorted_ar[:idx], depth + 1),
'right': build_kd_tree(sorted_ar[idx+1:], depth + 1)
}
def min_dist(p1, p2):
d1 = math.hypot(p1[0] - p2[0], p1[1] - p2[1])
return d1
# function designed to simultaneously recurse two trees to find the closest approach
def closest(k1,k2,lim=float("inf")):
cc1 = [k1[value] for value in k1 if k1[value] is not None and type(k1[value]) == dict]
cc2 = [k2[value] for value in k2 if k2[value] is not None and type(k2[value]) == dict]
if len(cc1) == 0 and len(cc2) == 0:
return min(lim, min_dist(k1['point'], k2['point']))
for md, c1, c2 in sorted((min_dist(c1['point'], c2['point']), c1, c2) for c1 in cc1 for c2 in cc2):
if md >= lim: break
lim = min(lim, closest(c1, c2, lim))
return lim
# some example coordinates
px_coords=np.array([299398.56,299402.16,299410.25,299419.7,299434.97,299443.75,299454.1,299465.3,299477.,299488.25,299496.8,299499.5,299501.28,299504.,299511.62,299520.62,299527.8,299530.06,299530.06,299525.12,299520.2,299513.88,299508.5,299500.84,299487.34,299474.78,299458.6,299444.66,299429.8,299415.4,299404.84,299399.47,299398.56,299398.56])
py_coords=np.array([822975.2,822989.56,823001.25,823005.3,823006.7,823005.06,823001.06,822993.4,822977.2,822961.,822943.94,822933.6,822925.06,822919.7,822916.94,822912.94,822906.6,822897.6,822886.8,822869.75,822860.75,822855.8,822855.4,822857.2,822863.44,822866.6,822870.6,822876.94,822886.8,822903.,822920.3,822937.44,822954.94,822975.2])
qx_coords=np.array([384072.1,384073.2,384078.9,384085.7,384092.47,384095.3,384097.12,384097.12,384093.9,384088.9,384082.47,384078.9,384076.03,384074.97,384073.53,384072.1])
qy_coords=np.array([780996.8,781001.1,781003.6,781003.6,780998.25,780993.25,780987.9,780981.8,780977.5,780974.7,780974.7,780977.2,780982.2,780988.25,780992.5,780996.8])
# some more example coordinates
#px_coords = np.array([299398,299402,299410.25,299419.7,299398])
#py_coords = np.array([822975.2,822920.3,822937.44,822954.94,822975.2])
#qx_coords = np.array([292316,292331.22,292329.72,292324.72,292319.44,292317.2,292316])
#qy_coords = np.array([663781,663788.25,663794,663798.06,663800.06,663799.3,663781])
# this is all just formatting the coordinates - only important thing to know is that p_midpoints and q_midpoints are two distinct sets of points, and are the targets in this question
px_edges = np.stack((px_coords, np.roll(px_coords, -1)),1)
px_midpoints = np.array(abs(px_coords + np.roll(px_coords, -1))/2)
py_edges = np.stack((py_coords, np.roll(py_coords, -1)),1)
py_midpoints = np.array(abs(py_coords + np.roll(py_coords, -1))/2)
p_edges = np.stack((px_edges, py_edges), axis=-1)[:-1]
p_midpoints = np.stack((px_midpoints, py_midpoints), axis=-1)[:-1]
qx_edges = np.stack((qx_coords, np.roll(qx_coords, -1)),1)
qx_midpoints = np.array(abs(qx_coords + np.roll(qx_coords, -1))/2)
qy_edges = np.stack((qy_coords, np.roll(qy_coords, -1)),1)
qy_midpoints = np.array(abs(qy_coords + np.roll(qy_coords, -1))/2)
q_edges = np.stack((qx_edges, qy_edges), axis=-1)[:-1]
q_midpoints = np.stack((qx_midpoints, qy_midpoints), axis=-1)[:-1]
# where the tree is actually built
p_tree = build_kd_tree(p_midpoints)
q_tree = build_kd_tree(q_midpoints)
# uncommect to see structure of tree
#pprint.pprint(p_tree)
near_distance = closest(p_tree, q_tree)
# brute force for testing
#distances = []
#for p_point in p_midpoints:
# for q_point in q_midpoints:
# distances.append(min_dist(p_point, q_point))
#
#m_dist = sorted(distances)[0]
#print(m_dist)
In my second attempt I tried to force the function to stop recursing when it hit the leaf of the tree. This works for the smaller of the two sample coordinate sets, but does not work for the larger of the two sample coordinate sets, failing with the same problem.
Second attempt - only 'closest' function, can be swapped out like-for-like with namesake in above code:
def closest(k1,k2,lim=float("inf")):
cc1 = [k1]
cc1 = cc1 + [k1[value] for value in k1 if k1[value] is not None and type(k1[value]) == dict]
cc2 = [k2]
cc2 = cc2 + [k2[value] for value in k2 if k2[value] is not None and type(k2[value]) == dict]
if len(cc1) == 1 and len(cc2) == 1:
return min(lim, min_dist(k1['point'], k2['point']))
md = [[min_dist(cc1[i]['point'], cc2[j]['point']), i, j, (cc1[i]['point'], cc2[j]['point'])] for i in range(len(cc1) >> 1, len(cc1)) for j in range(len(cc1) >> 1, len(cc2))]
md = sorted(md, key=itemgetter(0))
for h in range(0, len(md)):
lim = min(lim, closest(cc1[md[h][1]], cc2[md[h][2]],lim))
return lim
I'm aware that out-of-the-box solutions exist to solve this problem, but this is an area that I would like to understand better by building my own from scratch. Any help appreciated.