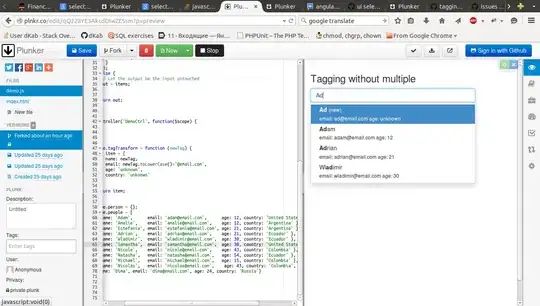
I am trying to color a sunburst by parent and then by level so that parent1 and parent2 are red and blue and then they gradient outwards by level.
I seem to be close using layout with sunburstcolorway but it doesn't quite color by level. Currently my input for colorway is a list of hex codes that are specified for each observation.
If there is a way to specify color for each individual section that would be ideal. I can't seem to figure it out, I'm very new to this
library(plotly)
clin2 <- data.frame(
stringsAsFactors = FALSE,
ids = c("MGH","CU","MGH - WT",
"CU - WT","MGH - KDM","CU - KDM","MGH - G2032R",
"MGH - S1986F","MGH - D2033N","CU - L1951R/L2026M"),
labels = c("Gainor<br>et al. 2017",
"McCoach<br>et al. 2018","ROS1<br>Extrinsic",
"ROS1<br>Extrinsic","ROS1<br>Intrinsic","ROS<br>Intrinsic","G2032R",
"S1986F","D2033N","L1951R/<br>L2026M"),
parents = c(NA,NA,"MGH","CU","MGH",
"CU","MGH - KDM","MGH - KDM","MGH - KDM","CU - KDM"),
values = c(17L, 12L, 8L, 11L, 9L, 1L, 7L, 1L, 1L, 1L),
colors = c("#3182bd", "#e6550d", "#9ecae1", "#fdae6b", "#9ecae1",
"#fdae6b", "#deebf7", "#deebf7", "#deebf7", "#fee6ce")
)
clin2_plot <- plot_ly(clin2, ids = ~ids, labels = ~labels, parents = ~parents,
values = ~values, type = 'sunburst', branchvalues = 'total'
) %>% layout(sunburstcolorway = clin2$colors)
clin2_plot
Thanks in advance.