I think there are 2 things that add confusion to this topic:
- statistical v.s. signal processing definition: as others have pointed out, in statistics we normalize auto-correlation into [-1,1].
- partial v.s. non-partial mean/variance: when the timeseries shifts at a lag>0, their overlap size will always < original length. Do we use the mean and std of the original (non-partial), or always compute a new mean and std using the ever changing overlap (partial) makes a difference. (There's probably a formal term for this, but I'm gonna use "partial" for now).
I've created 5 functions that compute auto-correlation of a 1d array, with partial v.s. non-partial distinctions. Some use formula from statistics, some use correlate in the signal processing sense, which can also be done via FFT. But all results are auto-correlations in the statistics definition, so they illustrate how they are linked to each other. Code below:
import numpy
import matplotlib.pyplot as plt
def autocorr1(x,lags):
'''numpy.corrcoef, partial'''
corr=[1. if l==0 else numpy.corrcoef(x[l:],x[:-l])[0][1] for l in lags]
return numpy.array(corr)
def autocorr2(x,lags):
'''manualy compute, non partial'''
mean=numpy.mean(x)
var=numpy.var(x)
xp=x-mean
corr=[1. if l==0 else numpy.sum(xp[l:]*xp[:-l])/len(x)/var for l in lags]
return numpy.array(corr)
def autocorr3(x,lags):
'''fft, pad 0s, non partial'''
n=len(x)
# pad 0s to 2n-1
ext_size=2*n-1
# nearest power of 2
fsize=2**numpy.ceil(numpy.log2(ext_size)).astype('int')
xp=x-numpy.mean(x)
var=numpy.var(x)
# do fft and ifft
cf=numpy.fft.fft(xp,fsize)
sf=cf.conjugate()*cf
corr=numpy.fft.ifft(sf).real
corr=corr/var/n
return corr[:len(lags)]
def autocorr4(x,lags):
'''fft, don't pad 0s, non partial'''
mean=x.mean()
var=numpy.var(x)
xp=x-mean
cf=numpy.fft.fft(xp)
sf=cf.conjugate()*cf
corr=numpy.fft.ifft(sf).real/var/len(x)
return corr[:len(lags)]
def autocorr5(x,lags):
'''numpy.correlate, non partial'''
mean=x.mean()
var=numpy.var(x)
xp=x-mean
corr=numpy.correlate(xp,xp,'full')[len(x)-1:]/var/len(x)
return corr[:len(lags)]
if __name__=='__main__':
y=[28,28,26,19,16,24,26,24,24,29,29,27,31,26,38,23,13,14,28,19,19,\
17,22,2,4,5,7,8,14,14,23]
y=numpy.array(y).astype('float')
lags=range(15)
fig,ax=plt.subplots()
for funcii, labelii in zip([autocorr1, autocorr2, autocorr3, autocorr4,
autocorr5], ['np.corrcoef, partial', 'manual, non-partial',
'fft, pad 0s, non-partial', 'fft, no padding, non-partial',
'np.correlate, non-partial']):
cii=funcii(y,lags)
print(labelii)
print(cii)
ax.plot(lags,cii,label=labelii)
ax.set_xlabel('lag')
ax.set_ylabel('correlation coefficient')
ax.legend()
plt.show()
Here is the output figure:
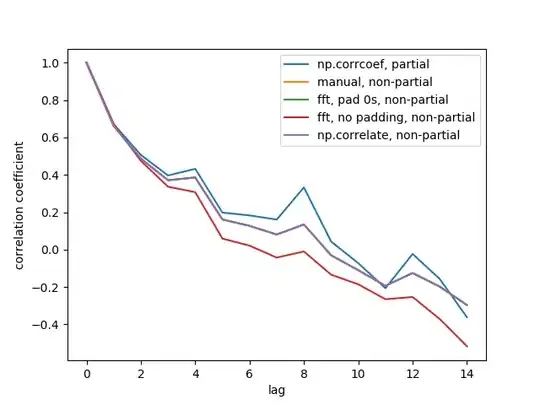
We don't see all 5 lines because 3 of them overlap (at the purple). The overlaps are all non-partial auto-correlations. This is because computations from the signal processing methods (np.correlate, FFT) don't compute a different mean/std for each overlap.
Also note that the fft, no padding, non-partial (red line) result is different, because it didn't pad the timeseries with 0s before doing FFT, so it's circular FFT. I can't explain in detail why, that's what I learned from elsewhere.
