Background: I picked up Python about a month ago, so my experience level is pretty slim. I'm pretty comfortable with VBA though years of data analysis through excel and PI Processbook.
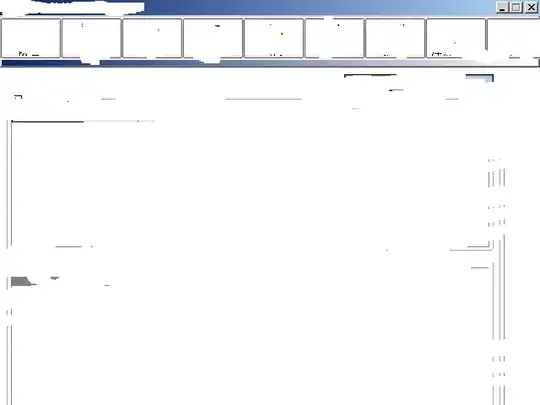
I have 27 thermocouples that I pull data for in 1s intervals. I would like to heatmap them from hottest to coldest at a given instance in time. I've leveraged seaborn heatmaps, but the problem with those is that they compare temperatures across time as well and the aggregate of these thermocouples changes dramatically over time. See chart below:
Notice how in the attached, the pink one is colder than the rest when all of them are cold, but when they all heat up, the cold spot transfers to the orange and green ones (and even the blue one for a little bit at the peak).
In excel, I would write a do loop to apply conditional formatting to each individual timestamp (row), however in Python I can't figure it out for the life of me. The following is the code that I used to develop the above chart, so I'm hoping I can modify this to make it work.
tsStartTime = pd.Timestamp(strStart_Time)
tsEndTime = pd.Timestamp(strEnd_Time)
t = np.linspace(tsStartTime.value,tsEndTime.value, 150301)
TimeAxis = pd.to_datetime(t)
fig,ax = plt.subplots(figsize=(25,5))
plt.subplots_adjust(bottom = 0.25)
x = TimeAxis
i = 1
while i < 28:
globals()['y' + str(i)] = forceconvert_v(globals()['arTTXD' + str(i)])
ax.plot(x,globals()['y' + str(i)])
i += 1
I've tried to use seaborn heatmaps, but when i slice it by timestamps, the output array is size (27,) instead of (27,1), so it gets rejected.
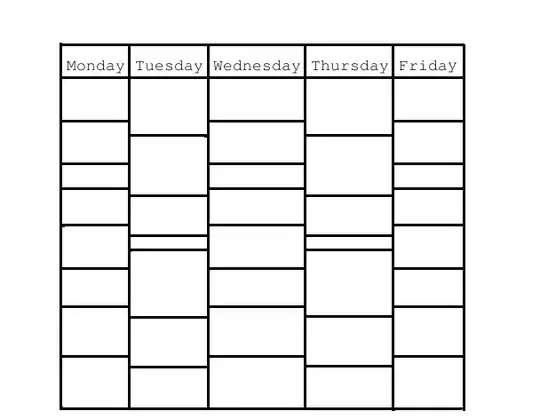
Ultimately, I'm looking for an output that looks like this:
Notice how the values of 15 in the middle are blue despite being higher than the red 5s in the beginning. I didnt fill out every cell, but hopefully you get the jist of what I'm trying to accomplish.
This data is being pulled from OSISoft PI via the PIConnect library. PI leverages their own classes, but they are essentially either series or dataframes, but I can manipulate them into whatever they need to be if someone has any awesome ideas to handle this.
Here's the link to the data: https://file.io/JS0RoQvDL6AB
Thanks!