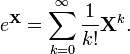If I run a time series box plot on an irregularly spaced time series (with missing values) with the date field as chr I get a box plot but with no gaps where the data is missing. If I import the data field as date and try the same plot I get the following error message:
Error in
geom_boxplot(): ! Problem while converting geom to grob. ℹ Error occurred in the 1st layer. Caused by error indraw_group(): ! Can only draw one boxplot per group ℹ Did you forgetaes(group = ...)?
I am not sure what variable I should be grouping on to get this to work. A sample of my data:
# A tibble: 17 × 9
SmplDate minDPM q1DPM medianDPM q3DPM maxDPM meanDPM stdevsDPM MeanBiomass
<date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2023-05-30 3 4.5 5.5 7.5 23.5 6.74 3.74 2.85
2 2005-06-02 6 16.4 20.5 25.5 52 21.9 9.87 7.56
3 2006-06-09 4 6.5 10.5 13.9 22.5 10.5 4.51 4.32
4 2007-06-07 4 7.5 10.5 13.9 36 11.5 5.67 4.65
5 2008-06-02 0.5 4 5 7 15.5 5.66 2.73 2.36
6 2009-06-01 2 6 8.25 11.9 23 9.39 4.23 3.91
7 2010-06-01 2.5 7.12 11 15.4 26.5 11.8 5.80 4.73
8 2011-05-30 1.5 4 5.25 8 18 6.6 3.88 2.79
9 2012-06-01 1.5 2.62 3.5 4.5 7.5 3.72 1.33 1.34
10 2013-06-05 2.5 5.12 6.25 8.5 17 7.01 2.73 2.96
11 2014-06-07 1.5 3 4 6.5 11.5 4.7 2.34 1.88
12 2016-05-31 3.5 8.25 12.5 16 59 13.3 8.11 5.21
13 2017-06-05 2 3.5 4 5 11.5 4.5 1.81 1.78
14 2018-06-08 1 4.62 6.25 9.88 28 7.78 5.09 3.29
15 2019-05-31 3 5.5 7 9 25 8.03 3.92 3.39
16 2021-06-05 3 6.5 8 12 24.5 9.41 4.42 3.91
17 2022-06-02 2 4.5 5.5 7.5 13 6.23 2.27 2.62
MY code so far:
# Time series boxplot
ggplot(data = vca01,
aes(x = SmplDate)) +
geom_boxplot(
aes(
ymin = minDPM,
lower = q1DPM,
middle = medianDPM,
upper = q3DPM,
ymax = maxDPM
),
stat = "identity") +
theme_bw() +
labs(x = "Year",
y = "Grass Biomass",
title = "Site name: VCA01")
I am wondering if this can be achieved if I converted the tibble to a zoo object? This is what I tried:
#convert tibble to zoo object
tseries <- read.zoo(vca01)
class(tseries)
# Time series Boxplot
ggplot(data = tseries,
aes(x = Index,
ymin = minDPM,
lower = q1DPM,
middle = medianDPM,
upper = q3DPM,
ymax = maxDPM)) +
geom_boxplot(stat = "identity") +
theme_bw() +
labs(x = "Year",
y = "Grass Biomass",
title = "Site name: VCA01")`
Error message:
Error in geom_boxplot():
! Problem while converting geom to grob.
ℹ Error occurred in the 1st layer.
Caused by error in draw_group():
! Can only draw one boxplot per group
ℹ Did you forget aes(group = ...)?
I tried the ggplot code with the data field as date and got the error described. I am not sure that melt (as suggested in R: Plot a time series with quantiles using ggplot2) will make any difference as my data are already in long format.