When the 2d-array (or nd-array) is C- or F-contiguous, then this task of mapping a function onto a 2d-array is practically the same as the task of mapping a function onto a 1d-array - we just have to view it that way, e.g. via np.ravel(A,'K').
Possible solution for 1d-array have been discussed for example here.
However, when the memory of the 2d-array isn't contiguous, then the situation a little bit more complicated, because one would like to avoid possible cache misses if axis are handled in wrong order.
Numpy has already a machinery in place to process axes in the best possible order. One possibility to use this machinery is np.vectorize. However, numpy's documentation on np.vectorize states that it is "provided primarily for convenience, not for performance" - a slow python function stays a slow python function with the whole associated overhead! Another issue is its huge memory-consumption - see for example this SO-post.
When one wants to have a performance of a C-function but to use numpy's machinery, a good solution is to use numba for creation of ufuncs, for example:
# runtime generated C-function as ufunc
import numba as nb
@nb.vectorize(target="cpu")
def nb_vf(x):
return x+2*x*x+4*x*x*x
It easily beats np.vectorize but also when the same function would be performed as numpy-array multiplication/addition, i.e.
# numpy-functionality
def f(x):
return x+2*x*x+4*x*x*x
# python-function as ufunc
import numpy as np
vf=np.vectorize(f)
vf.__name__="vf"
See appendix of this answer for time-measurement-code:
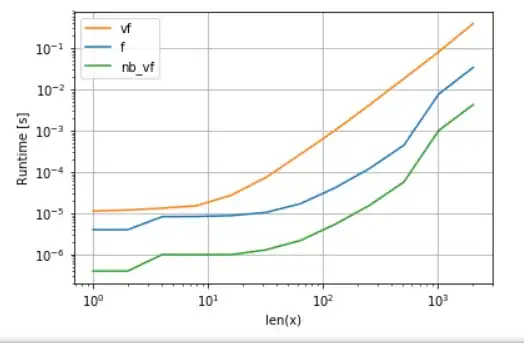
Numba's version (green) is about 100 times faster than the python-function (i.e. np.vectorize), which is not surprising. But it is also about 10 times faster than the numpy-functionality, because numbas version doesn't need intermediate arrays and thus uses cache more efficiently.
While numba's ufunc approach is a good trade-off between usability and performance, it is still not the best we can do. Yet there is no silver bullet or an approach best for any task - one has to understand what are the limitation and how they can be mitigated.
For example, for transcendental functions (e.g. exp, sin, cos) numba doesn't provide any advantages over numpy's np.exp (there are no temporary arrays created - the main source of the speed-up). However, my Anaconda installation utilizes Intel's VML for vectors bigger than 8192 - it just cannot do it if memory is not contiguous. So it might be better to copy the elements to a contiguous memory in order to be able to use Intel's VML:
import numba as nb
@nb.vectorize(target="cpu")
def nb_vexp(x):
return np.exp(x)
def np_copy_exp(x):
copy = np.ravel(x, 'K')
return np.exp(copy).reshape(x.shape)
For the fairness of the comparison, I have switched off VML's parallelization (see code in the appendix):
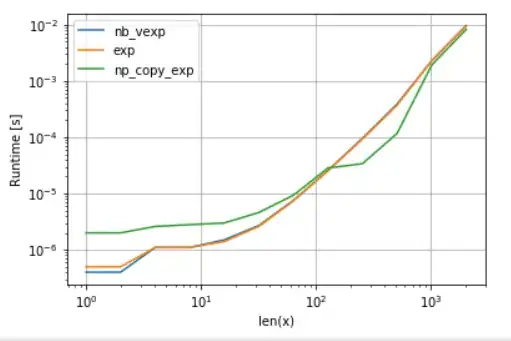
As one can see, once VML kicks in, the overhead of copying is more than compensated. Yet once data becomes too big for L3 cache, the advantage is minimal as task becomes once again memory-bandwidth-bound.
On the other hand, numba could use Intel's SVML as well, as explained in this post:
from llvmlite import binding
# set before import
binding.set_option('SVML', '-vector-library=SVML')
import numba as nb
@nb.vectorize(target="cpu")
def nb_vexp_svml(x):
return np.exp(x)
and using VML with parallelization yields:
numba's version has less overhead, but for some sizes VML beats SVML even despite of the additional copying overhead - which isn't a bit surprise as numba's ufuncs aren't parallelized.
Listings:
A. comparison of polynomial function:
import perfplot
perfplot.show(
setup=lambda n: np.random.rand(n,n)[::2,::2],
n_range=[2**k for k in range(0,12)],
kernels=[
f,
vf,
nb_vf
],
logx=True,
logy=True,
xlabel='len(x)'
)
B. comparison of exp:
import perfplot
import numexpr as ne # using ne is the easiest way to set vml_num_threads
ne.set_vml_num_threads(1)
perfplot.show(
setup=lambda n: np.random.rand(n,n)[::2,::2],
n_range=[2**k for k in range(0,12)],
kernels=[
nb_vexp,
np.exp,
np_copy_exp,
],
logx=True,
logy=True,
xlabel='len(x)',
)


