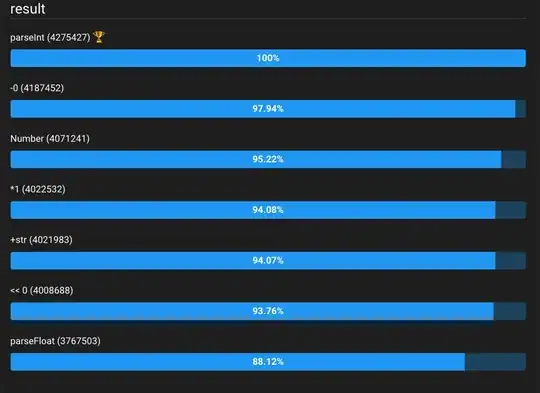
Please download the data here!
Target: Plot an image like this:
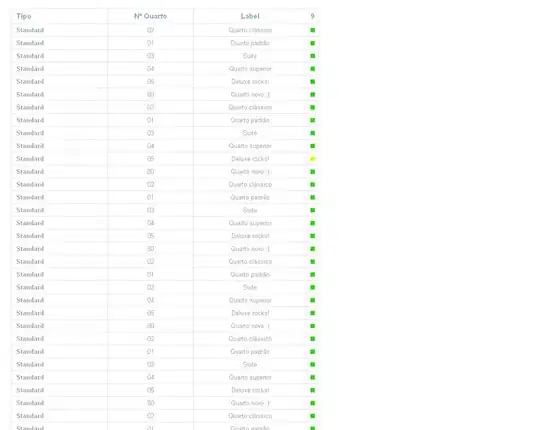
Features: 1. two different time series; 2. the lower panel has a reverse y-axis; 3. shadows over two plots.
Possible solutions:
1. Facetting is not appropriate - (1) can not just make one facet's y axis reverse while keep the other(s) unchanges. (2) difficult to adjust the individual facets one by one.
2. Using viewports to arrange individual plots using the following codes:
library(ggplot2)
library(grid)
library(gridExtra)
##Import data
df<- read.csv("D:\\R\\SF_Question.csv")
##Draw individual plots
#the lower panel
p1<- ggplot(df, aes(TIME1, VARIABLE1)) + geom_line() + scale_y_reverse() + labs(x="AGE") + scale_x_continuous(breaks = seq(1000,2000,200), limits = c(1000,2000))
#the upper panel
p2<- ggplot(df, aes(TIME2, V2)) + geom_line() + labs(x=NULL) + scale_x_continuous(breaks = seq(1000,2000,200), limits = c(1000,2000)) + theme(axis.text.x=element_blank())
##For the shadows
#shadow position
rects<- data.frame(x1=c(1100,1800),x2=c(1300,1850),y1=c(0,0),y2=c(100,100))
#make shadows clean (hide axis, ticks, labels, background and grids)
xquiet <- scale_x_continuous("", breaks = NULL)
yquiet <- scale_y_continuous("", breaks = NULL)
bgquiet<- theme(panel.background = element_rect(fill = "transparent", colour = NA))
plotquiet<- theme(plot.background = element_rect(fill = "transparent", colour = NA))
quiet <- list(xquiet, yquiet, bgquiet, plotquiet)
prects<- ggplot(rects,aes(xmin=x1,xmax=x2,ymin=y1,ymax=y2))+ geom_rect(alpha=0.1,fill="blue") + coord_cartesian(xlim = c(1000, 2000)) + quiet
##Arrange plots
pushViewport(viewport(layout = grid.layout(2, 1)))
vplayout <- function(x, y)
viewport(layout.pos.row = x, layout.pos.col = y)
#arrange time series
print(p2, vp = vplayout(1, 1))
print(p1, vp = vplayout(2, 1))
#arrange shadows
print(prects, vp=vplayout(1:2,1))
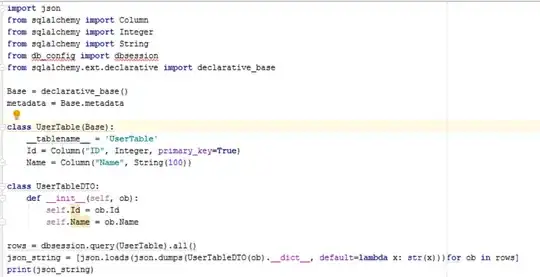
Problems:
- the x-axis doesn't align correctly;
- the shadow locations are wrong (because of the incorrect line-up of x-axis).
After Googling all around:
- I firstly noticed that "align.plots() from ggExtra" could do this job. However, it has been deprecated by the author;
- Then I've tried the gglayout solution, but no luck - I even could not install the "cutting-edge" package;
Finally, I've tried the gtable solution using the following code:
gp1<- ggplot_gtable(ggplot_build(p1)) gp2<- ggplot_gtable(ggplot_build(p2)) gprects<- ggplot_gtable(ggplot_build(prects)) maxWidth = unit.pmax(gp1$widths[2:3], gp2$widths[2:3], gprects$widths[2:3]) gp1$widths[2:3] <- maxWidth gp2$widths[2:3] <- maxWidth gprects$widths[2:3] <- maxWidth grid.arrange(gp2, gp1, gprects)

Now, the x-axis of upper and lower panel do align correctly. But the shadow positions are still wrong. And more importantly, I can not overlap the shadow plot on the two time-series. After several day's attempts, I nearly give up...
Could somebody here give me a hand?