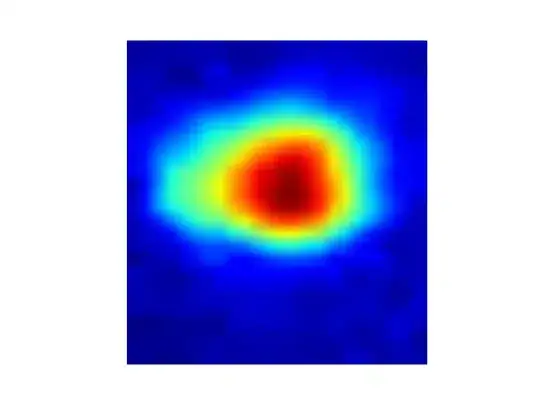
I have a series of basic 2D images (3 for simplicity for now) and these are related to each other, analogous to frames from a movie:
Within python how may I stack these slices on top of each other, as in image1->image2->image-3? I'm using pylab to display these images. Ideally an isometric view of the stacked frames would be good or a tool allowing me to rotate the view within code/in rendered image.
Any assistance appreciated. Code and images shown:
from PIL import Image
import pylab
fileName = "image1.png"
im = Image.open(fileName)
pylab.axis('off')
pylab.imshow(im)
pylab.show()