converters or dtype won't always help. Especially for date/time and duration (ideally a mix of both...), post-processing is necessary. In such cases, reading the Excel file's content to a built-in type and create the DataFrame from that can be an option.
Here's an example file. The "duration" column contains duration values in HH:MM:SS and invalid values "-".
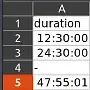
If the hour is less than 24, Excel formats the entry to a time, otherwise a duration. We want dtype timedelta for the whole column in the DataFrame. But pandas messes up the import:
import pandas as pd
df = pd.read_excel("path-to-file")
df.duration
# 0 12:30:00
# 1 1900-01-01 00:30:00
# 2 -
# 3 1900-01-01 23:55:01
# Name: duration, dtype: object
[type(i) for i in df.duration]
# [datetime.time, datetime.datetime, str, datetime.datetime]
Now we have datetime.datetime and datetime.time objects, and it's difficult to get back duration (timedelta)! You could do it directly with a converter, but that does not make it less difficult.
Here, I found it to be actually easier to use the excel loader engine directly:
from openpyxl import load_workbook
wb = load_workbook('path-to-file')
sheet = wb['Tests'] # adjust sheet name, this is for the demo file
data = list(sheet.values) # a list of tuples, one tuple for each row
df = pd.DataFrame(data[1:], columns=data[0]) # first tuple is column names
df['duration']
# 0 12:30:00
# 1 1 day, 0:30:00
# 2 -
# 3 1 day, 23:55:01
# Name: duration, dtype: object
[type(i) for i in df['duration']]
# [datetime.time, datetime.timedelta, str, datetime.timedelta]
So now we already have some timedelta objects! The conversion of the others to timedelta can be done as simple as
df['duration'] = pd.to_timedelta(df.duration.astype(str), errors='coerce')
df['duration']
# 0 0 days 12:30:00
# 1 1 days 00:30:00
# 2 NaT
# 3 1 days 23:55:01
# Name: duration, dtype: timedelta64[ns]
