We are using the registration algorithm of ITK but we only want the affine transformation matrix and not directly apply the registration. In a previous issues we already solved a misunderstanding regarding the image/transform orientation: How to get transformation affine from ITK registration?
We did now run into a sample where the current solution does not properly work. The rotation is good but the result is slightly translated. The image output of ITK is perfect, so we know that the registration worked. That's why we will reduce the problem description below to the affine calculation with the specific matrices.
From the ITK registration we get/read the following parameters:
parameter_map = result_transform_parameters.GetParameterMap(0)
rot00, rot01, rot02, rot10, rot11, rot12, rot20, rot21, rot22 = parameter_map[
'TransformParameters'][:9]
A = np.array([
[rot00, rot01, rot02, 0],
[rot10, rot11, rot12, 0],
[rot20, rot21, rot22, 0],
[ 0, 0, 0, 1],
], dtype=float) # yapf: disable
tx, ty, tz = parameter_map['TransformParameters'][9:]
t = np.array([
[1, 0, 0, tx],
[0, 1, 0, ty],
[0, 0, 1, tz],
[0, 0, 0, 1],
], dtype=float) # yapf: disable
# In world coordinates
cx, cy, cz = parameter_map['CenterOfRotationPoint']
c = np.array([
[1, 0, 0, cx],
[0, 1, 0, cy],
[0, 0, 1, cz],
[0, 0, 0, 1],
], dtype=float) # yapf: disable
ox, oy, oz = parameter_map['Origin']
o = np.array([
[1, 0, 0, ox],
[0, 1, 0, oy],
[0, 0, 1, oz],
[0, 0, 0, 1],
], dtype=float) # yapf: disable
moving_ras = moving_image.affine
Where A is the direction/rotation matrix, t the translation matrix, c the center of rotation (CoR), and moving_ras the affine of the moving image in RAS orientation.
The translation and direction matrix can be combined to one transform matrix:
transform = t @ A
We are not sure how to factor in the CenterOfRotationPoint.
Based on this, this, and that exchange questions, I thought one might need to do it like that:
transform = c @ transform @ np.linalg.inv(c)
Finally, we need to add the orientation flip between RAS and LPS:
registration = FLIPXY_44 @ transform @ FLIPXY_44
But this does not result in the correct transformation affine.
On the ITK docs and in a GitHub issue we got this formula to apply the above parameters to points:
T(x) = A ( x - c ) + (t + c)
While we can not directly use that since we do not want to directly transform the image but we only want to calculate the correct affine transformation matrix, one can see how the formula is pretty similar to what we are already doing as explained above.
We are again at a dead end with our knowledge.
Things we noticed that might make issues here:
- Orientation
- ITK uses LPS orientation for images and transforms
- Monai/Nibabel uses RAS orientation for images and transforms
- Center of Rotation
- ITK provides the used center of rotation
- Monai implicitly assumes the center of rotation to be the center of the image
- World space vs. Index space.
- All transforms and points from ITK are in world space.
- Monai seems to operate directly on the image.
- (0, 0, 0) Corner - ITK and Monai seem to use the opposit corner for coordinates - e.g. in a 4x4x4 image, position (0, 0, 0) in ITK is position (3, 3, 3) in Monai.
EDIT: I noticed that my current minimal code example is not quite comprehensive. Therefore here an update. The included affine matrices are taken from the ITK coregistration. The ITK code was omitted for brevity.
Here with new test data (you can view these images via MRIcoGL):
Here a minimal code example:
from pathlib import Path
import nibabel
import numpy as np
from monai.transforms.spatial.array import Affine
from monai.utils.enums import GridSampleMode, GridSamplePadMode
from nibabel import Nifti1Image
np.set_printoptions(suppress=True) # type: ignore
folder = Path('.')
FLIPXY_44 = np.diag([-1, -1, 1, 1])
# rot00, rot01, rot02, rot10, rot11, rot12, rot20, rot21, rot22 = parameter_map['TransformParameters'][:9]
A = np.array([[ 1.02380734, -0.05137566, -0.00766465, 0. ],
[ 0.01916231, 0.93276486, -0.23453097, 0. ],
[ 0.01808809, 0.2667324 , 0.94271694, 0. ],
[ 0. , 0. , 0. , 1. ]]) # yapf: disable
# tx, ty, tz = parameter_map['TransformParameters'][9:]
t = np.array([[ 1. , 0. , 0. , 1.12915465 ],
[ 0. , 1. , 0. , 11.76880151 ],
[ 0. , 0. , 1. , 41.54685788 ],
[ 0. , 0. , 0. , 1. ]]) # yapf: disable
# cx, cy, cz = parameter_map['CenterOfRotationPoint']
c = np.array([[ 1. , 0. , 0. , -0.1015625 ],
[ 0. , 1. , 0. , -24.5521698 ],
[ 0. , 0. , 1. , 0.1015625 ],
[ 0. , 0. , 0. , 1. ]]) # yapf: disable
# Moving image affine
x = np.array([[ 2. , 0. , 0. , -125.75732422],
[ 0. , 2. , 0. , -125.23828888],
[ 0. , 0. , 2. , -99.86506653],
[ 0. , 0. , 0. , 1. ]]) # yapf: disable
o = np.array([
[1., 0., 0., 126.8984375],
[0., 1., 0., 102.4478302],
[0., 0., 1., -126.8984375],
[0., 0., 0., 1.],
])
moving_ras = x
# Combine the direction and translation
transform = t @ A
# Factor in the center of rotation
# transform = c @ transform @ np.linalg.inv(c)
# Switch from LPS to RAS orientation
registration = FLIPXY_44 @ transform @ FLIPXY_44
y = np.array([[ 2. , 0. , 0. , -126.8984375 ],
[ 0. , 2. , 0. , -102.4478302 ],
[ 0. , 0. , 2. , -126.8984375 ],
[ 0. , 0. , 0. , 1. ]]) # yapf: disable
fixed_image_affine = y
moving_image_ni: Nifti1Image = nibabel.load(folder / 'real_moving.nii.gz')
moving_image_np: np.ndarray = moving_image_ni.get_fdata() # type: ignore
affine_transform = Affine(affine=registration,
image_only=True,
mode=GridSampleMode.NEAREST,
padding_mode=GridSamplePadMode.BORDER)
reg_monai = np.squeeze(affine_transform(moving_image_np[np.newaxis, ...]))
out = Nifti1Image(reg_monai, fixed_image_affine)
nibabel.save(out, folder / 'reg_monai.nii.gz')
When you executed this code, the resulting reg_monai.nii.gz should match the real_fixed.nii.gz (in position and outline - not in the actual content).
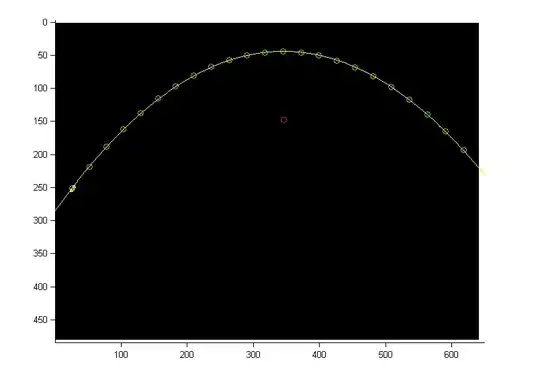
Currently the result looks like this (viewed via MRIcoGL):
But the result should look like this (this is the direct ITK registration output where the hardcoded affine matrices come from - which should prove that the registration worked and that the parameters generally should be good):
For the sake of completeness, here also the code to perform the ITK registration and to get the above affine matrices:
from pathlib import Path
import itk
import numpy as np
np.set_printoptions(suppress=True) # type: ignore
folder = Path('.')
moving_image = itk.imread(str(folder / 'real_moving.nii.gz'), itk.F)
fixed_image = itk.imread(str(folder / 'real_fixed.nii.gz'), itk.F)
# Import Default Parameter Map
parameter_object = itk.ParameterObject.New()
affine_parameter_map = parameter_object.GetDefaultParameterMap('affine', 4)
affine_parameter_map['FinalBSplineInterpolationOrder'] = ['1']
affine_parameter_map['MaximumNumberOfIterations'] = ['512']
parameter_object.AddParameterMap(affine_parameter_map)
# Call registration function
result_image, result_transform_parameters = itk.elastix_registration_method( # type: ignore
fixed_image, moving_image, parameter_object=parameter_object)
itk.imwrite(result_image, str(folder / 'real_reg_itk.nii.gz'), compression=True)
parameter_map = result_transform_parameters.GetParameterMap(0)
rot00, rot01, rot02, rot10, rot11, rot12, rot20, rot21, rot22 = parameter_map['TransformParameters'][:9]
A = np.array([
[rot00, rot01, rot02, 0],
[rot10, rot11, rot12, 0],
[rot20, rot21, rot22, 0],
[ 0, 0, 0, 1],
], dtype=float) # yapf: disable
tx, ty, tz = parameter_map['TransformParameters'][9:]
t = np.array([
[1, 0, 0, tx],
[0, 1, 0, ty],
[0, 0, 1, tz],
[0, 0, 0, 1],
], dtype=float) # yapf: disable
# In world coordinates
cx, cy, cz = parameter_map['CenterOfRotationPoint']
c = np.array([
[1, 0, 0, cx],
[0, 1, 0, cy],
[0, 0, 1, cz],
[0, 0, 0, 1],
], dtype=float) # yapf: disable
ox, oy, oz = parameter_map['Origin']
o = np.array([
[1, 0, 0, ox],
[0, 1, 0, oy],
[0, 0, 1, oz],
[0, 0, 0, 1],
], dtype=float) # yapf: disable
Package versions:
itk-elastix==0.12.0
monai==0.8.0
nibabel==3.1.1
numpy==1.19.2