I'm creating a facetted plot to view predicted vs. actual values side by side with a plot of predicted value vs. residuals. I'll be using shiny to help explore the results of modeling efforts using different training parameters. I train the model with 85% of the data, test on the remaining 15%, and repeat this 5 times, collecting actual/predicted values each time. After calculating the residuals, my data.frame looks like this:
head(results)
act pred resid
2 52.81000 52.86750 -0.05750133
3 44.46000 42.76825 1.69175252
4 54.58667 49.00482 5.58184181
5 36.23333 35.52386 0.70947731
6 53.22667 48.79429 4.43237981
7 41.72333 41.57504 0.14829173
What I want:
- Side by side plot of
predvs.actandpredvs.resid - The x/y range/limits for
predvs.actto be the same, ideally frommin(min(results$act), min(results$pred))tomax(max(results$act), max(results$pred)) - The x/y range/limits for
predvs.residnot to be affected by what I do to the actual vs. predicted plot. Plotting forxover only the predicted values andyover only the residual range is fine.
In order to view both plots side by side, I melt the data:
library(reshape2)
plot <- melt(results, id.vars = "pred")
Now plot:
library(ggplot2)
p <- ggplot(plot, aes(x = pred, y = value)) + geom_point(size = 2.5) + theme_bw()
p <- p + facet_wrap(~variable, scales = "free")
print(p)
That's pretty close to what I want:
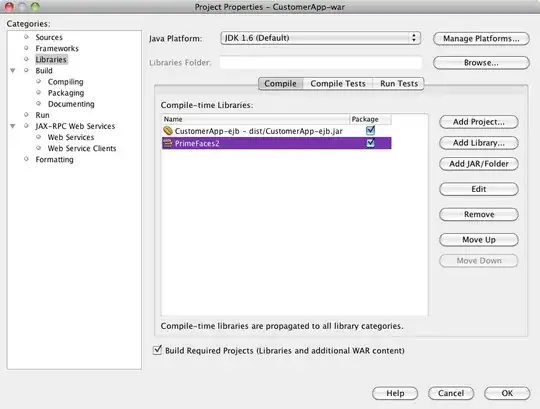
What I'd like is for the x and y ranges for actual vs. predicted to be the same, but I'm not sure how to specify that, and I don't need that done for the predicted vs. residual plot since the ranges are completely different.
I tried adding something like this for both scale_x_continous and scale_y_continuous:
min_xy <- min(min(plot$pred), min(plot$value))
max_xy <- max(max(plot$pred), max(plot$value))
p <- ggplot(plot, aes(x = pred, y = value)) + geom_point(size = 2.5) + theme_bw()
p <- p + facet_wrap(~variable, scales = "free")
p <- p + scale_x_continuous(limits = c(min_xy, max_xy))
p <- p + scale_y_continuous(limits = c(min_xy, max_xy))
print(p)
But that picks up the min() of the residual values.
One last idea I had is to store the value of the minimum act and pred variables before melting, and then add them to the melted data frame in order to dictate in which facet they appear:
head(results)
act pred resid
2 52.81000 52.86750 -0.05750133
3 44.46000 42.76825 1.69175252
4 54.58667 49.00482 5.58184181
5 36.23333 35.52386 0.70947731
min_xy <- min(min(results$act), min(results$pred))
max_xy <- max(max(results$act), max(results$pred))
plot <- melt(results, id.vars = "pred")
plot <- rbind(plot, data.frame(pred = c(min_xy, max_xy),
variable = c("act", "act"), value = c(max_xy, min_xy)))
p <- ggplot(plot, aes(x = pred, y = value)) + geom_point(size = 2.5) + theme_bw()
p <- p + facet_wrap(~variable, scales = "free")
print(p)
That does what I want, with the exception that the points show up, too:

Any suggestions for doing something like this?
I saw this idea to add geom_blank(), but I'm not sure how to specify the aes() bit and have it work properly, or what the geom_point() equivalent is to the histogram use of aes(y = max(..count..)).
Here's data to play with (my actual, predicted, and residual values prior to melting):
results <- read.table(header = TRUE, text = "
act pred resid
52.81 52.8675013282404 -0.0575013282403773
44.46 42.7682474758679 1.69175252413213
54.5866666666667 49.0048248585123 5.58184180815435
36.2333333333333 35.5238560262515 0.709477307081826
53.2266666666667 48.7942868566949 4.43237980997177
41.7233333333333 41.5750416040131 0.148291729320228
35.2966666666667 33.9548164913007 1.34185017536599
30.6833333333333 29.9787449128663 0.704588420467079
39.25 37.6443975781139 1.60560242188613
35.8866666666667 36.7196211666685 -0.832954500001826
25.1 27.6043278172077 -2.50432781720766
29.0466666666667 27.0615724310721 1.98509423559461
23.2766666666667 31.2073056885252 -7.93063902185855
56.3866666666667 55.0886903524179 1.29797631424874
42.92 43.0895814712768 -0.169581471276786
41.57 43.0895814712768 -1.51958147127679
27.92 32.3549865881578 -4.43498658815778
23.16 26.2428426737583 -3.08284267375831
38.0166666666667 36.6926037128343 1.32406295383237
61.8966666666667 56.7987490221996 5.09791764446704
37.41 45.0370788180147 -7.62707881801468
41.6333333333333 41.8231642271826 -0.189830893849219
35.9466666666667 38.3297859332601 -2.38311926659339
48.9933333333333 49.5343916620086 -0.541058328675241
30.5666666666667 30.8535641206809 -0.286897454014273
32.08 29.0117492750411 3.06825072495888
40.3633333333333 36.9767968381391 3.38653649519422
53.2266666666667 49.0826677983065 4.14399886836018
64.6066666666667 54.4678549541069 10.1388117125598
38.5366666666667 35.5059204731218 3.03074619354486
41.7233333333333 41.5333417555995 0.189991577733821
25.78 27.6069075391361 -1.82690753913609
33.4066666666667 31.2404889715121 2.16617769515461
27.8033333333333 27.8920960978598 -0.088762764526507
39.3266666666667 37.8505531149324 1.47611355173427
48.9933333333333 49.2616631533957 -0.268329820062384
25.2433333333333 30.366837650159 -5.12350431682565
32.67 31.1623492639066 1.5076507360934
55.17 55.0456078770405 0.124392122959534
42.92 42.772538591063 0.147461408936991
54.5866666666667 49.2419293590535 5.34473730761318
23.16 26.1963523976241 -3.03635239762411
64.6066666666667 54.4080781796616 10.1985884870051
40.7966666666667 44.9796700541254 -4.18300338745873
39.0166666666667 34.6996927469131 4.31697391975358
41.6333333333333 41.6227713664027 0.0105619669306023
35.8866666666667 36.8449646519306 -0.958297985263961
25.1 27.5318686661673 -2.43186866616734
23.2766666666667 31.6641793552795 -8.38751268861282
44.46 42.8198894266632 1.64011057333683
34.2166666666667 40.5769177148146 -6.36025104814794
40.8033333333333 40.5769177148146 0.226415618518729
24.5766666666667 29.3807781312816 -4.80411146461488
35.73 36.8579132935989 -1.1279132935989
61.8966666666667 55.5617033901752 6.33496327649151
62.1833333333333 55.8097119335638 6.37362139976954
74.6466666666667 55.1041728261666 19.5424938405001
39.4366666666667 43.6094641699075 -4.17279750324084
36.6 37.0674887276681 -0.467488727668119
27.1333333333333 27.3876960746536 -0.254362741320246
")