Before answering the actual question we should ask another one that's very relevant depending on the nature of your data:
What is an outlier?
Imagine the series of values [3, 2, 3, 4, 999] (where the 999 seemingly doesn't fit in) and analyse various ways of outlier detection
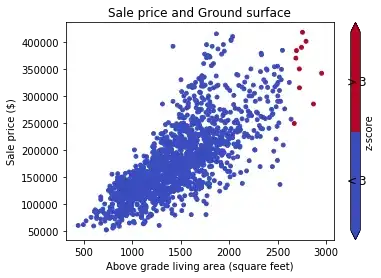
Z-Score
The problem here is that the value in question distorts our measures mean and std heavily, resulting in inconspicious z-scores of roughly [-0.5, -0.5, -0.5, -0.5, 2.0], keeping every value within two standard deviations of the mean. One very large outlier might hence distort your whole assessment of outliers. I would discourage this approach.
Quantile Filter
A way more robust approach is given is this answer, eliminating the bottom and top 1% of data. However, this eliminates a fixed fraction independant of the question if these data are really outliers. You might loose a lot of valid data, and on the other hand still keep some outliers if you have more than 1% or 2% of your data as outliers.
IQR-distance from Median
Even more robust version of the quantile principle: Eliminate all data that is more than f times the interquartile range away from the median of the data. That's also the transformation that sklearn's RobustScaler uses for example. IQR and median are robust to outliers, so you outsmart the problems of the z-score approach.
In a normal distribution, we have roughly iqr=1.35*s, so you would translate z=3 of a z-score filter to f=2.22 of an iqr-filter. This will drop the 999 in the above example.
The basic assumption is that at least the "middle half" of your data is valid and resembles the distribution well, whereas you also mess up if your distribution has wide tails and a narrow q_25% to q_75% interval.
Advanced Statistical Methods
Of course there are fancy mathematical methods like the Peirce criterion, Grubb's test or Dixon's Q-test just to mention a few that are also suitable for non-normally distributed data. None of them are easily implemented and hence not addressed further.
Code
Replacing all outliers for all numerical columns with np.nan on an example data frame. The method is robust against all dtypes that pandas provides and can easily be applied to data frames with mixed types:
import pandas as pd
import numpy as np
# sample data of all dtypes in pandas (column 'a' has an outlier) # dtype:
df = pd.DataFrame({'a': list(np.random.rand(8)) + [123456, np.nan], # float64
'b': [0,1,2,3,np.nan,5,6,np.nan,8,9], # int64
'c': [np.nan] + list("qwertzuio"), # object
'd': [pd.to_datetime(_) for _ in range(10)], # datetime64[ns]
'e': [pd.Timedelta(_) for _ in range(10)], # timedelta[ns]
'f': [True] * 5 + [False] * 5, # bool
'g': pd.Series(list("abcbabbcaa"), dtype="category")}) # category
cols = df.select_dtypes('number').columns # limits to a (float), b (int) and e (timedelta)
df_sub = df.loc[:, cols]
# OPTION 1: z-score filter: z-score < 3
lim = np.abs((df_sub - df_sub.mean()) / df_sub.std(ddof=0)) < 3
# OPTION 2: quantile filter: discard 1% upper / lower values
lim = np.logical_and(df_sub < df_sub.quantile(0.99, numeric_only=False),
df_sub > df_sub.quantile(0.01, numeric_only=False))
# OPTION 3: iqr filter: within 2.22 IQR (equiv. to z-score < 3)
iqr = df_sub.quantile(0.75, numeric_only=False) - df_sub.quantile(0.25, numeric_only=False)
lim = np.abs((df_sub - df_sub.median()) / iqr) < 2.22
# replace outliers with nan
df.loc[:, cols] = df_sub.where(lim, np.nan)
To drop all rows that contain at least one nan-value:
df.dropna(subset=cols, inplace=True) # drop rows with NaN in numerical columns
# or
df.dropna(inplace=True) # drop rows with NaN in any column
Using pandas 1.3 functions: